bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_6025_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
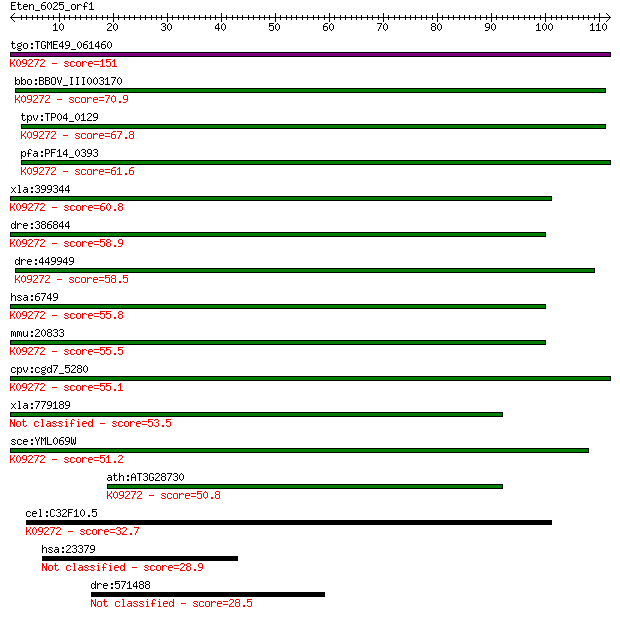
tgo:TGME49_061460 structure specific recognition protein I, pu... 151 4e-37
bbo:BBOV_III003170 17.m07304; structure specific recognition p... 70.9 9e-13
tpv:TP04_0129 structure specific recognition protein; K09272 s... 67.8 7e-12
pfa:PF14_0393 structure specific recognition protein; K09272 s... 61.6 6e-10
xla:399344 ssrp1; structure specific recognition protein 1; K0... 60.8 1e-09
dre:386844 ssrp1a, ssrp1, wu:fa11b03, wu:fd10f06; structure sp... 58.9 4e-09
dre:449949 ssrp1b, im:7140964; structure specific recognition ... 58.5 5e-09
hsa:6749 SSRP1, FACT, FACT80, T160; structure specific recogni... 55.8 3e-08
mmu:20833 Ssrp1, C81323, Hmg1-rs1, Hmgi-rs3, Hmgox, T160; stru... 55.5 4e-08
cpv:cgd7_5280 structure-specific recognition protein 1 (SSRP1)... 55.1 5e-08
xla:779189 raph1, MGC114656; Ras association (RalGDS/AF-6) and... 53.5 1e-07
sce:YML069W POB3; Pob3p; K09272 structure-specific recognition... 51.2 7e-07
ath:AT3G28730 ATHMG; ATHMG (ARABIDOPSIS THALIANA HIGH MOBILITY... 50.8 1e-06
cel:C32F10.5 hmg-3; HMG family member (hmg-3); K09272 structur... 32.7 0.26
hsa:23379 DKFZp781J2344; KIAA0947 28.9 4.7
dre:571488 tripartite motif protein TRIM29-like 28.5 5.5
> tgo:TGME49_061460 structure specific recognition protein I,
putative ; K09272 structure-specific recognition protein 1
Length=539
Score = 151 bits (382), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 68/111 (61%), Positives = 87/111 (78%), Gaps = 0/111 (0%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
+ D+ RHF HFK++LQ A+ HWGDV M+GNNL ++V+GC AF+++AQ+IAQVTT
Sbjct 106 FADLSRHFDAHFKVKLQRGQQAYRGWHWGDVKMEGNNLQLTVDGCAAFDIHAQEIAQVTT 165
Query 61 PSKNDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDETDSHLQNLKEKLVK 111
PSKNDLAIEL+QDDT DQ ED LLE+R +QP GDD+ + LQ LK+KLVK
Sbjct 166 PSKNDLAIELIQDDTRDQQEDQLLEVRFYQPFAGDDDAEGPLQQLKQKLVK 216
> bbo:BBOV_III003170 17.m07304; structure specific recognition
protein; K09272 structure-specific recognition protein 1
Length=485
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 33/110 (30%), Positives = 64/110 (58%), Gaps = 2/110 (1%)
Query 2 EDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTP 61
+++++HF +HFK+ + ++AH HWG + + ++++ +++A+D+ QVT P
Sbjct 88 DELKQHFDEHFKISPEVGSVAHTGWHWGVYGFENDTFKLTIDDNAGIDIDAKDVTQVTVP 147
Query 62 SKNDLAIELVQDDTMDQSEDMLLEIRLFQPATGD-DETDSHLQNLKEKLV 110
+K DLA+E Q+ D L+EIR P D D+ + L++LK+ +
Sbjct 148 TKTDLAVEFKQNKGYING-DELMEIRFCIPNKPDVDDNELALEDLKQTFL 196
> tpv:TP04_0129 structure specific recognition protein; K09272
structure-specific recognition protein 1
Length=460
Score = 67.8 bits (164), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 36/109 (33%), Positives = 59/109 (54%), Gaps = 2/109 (1%)
Query 3 DIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPS 62
D+ +HF +++K+ + ++ HWG D + +N +++AQ I Q T PS
Sbjct 108 DLSKHFEENYKMSCEKDEVSCTGWHWGTYEFDNTTFRLRINNNSGLDIDAQSIIQATIPS 167
Query 63 KNDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDE-TDSHLQNLKEKLV 110
K DLAIEL +T++ S D L+EIR P D E + L++LK+ +
Sbjct 168 KTDLAIELKNVNTLNNS-DELVEIRFCLPNKLDPEDNEIQLEDLKQTFL 215
> pfa:PF14_0393 structure specific recognition protein; K09272
structure-specific recognition protein 1
Length=506
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 61/110 (55%), Gaps = 3/110 (2%)
Query 3 DIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPS 62
+I +HF +F ++L + IA +WG+ ++ +NL ++ AF + +I Q+
Sbjct 95 EITQHFQKYFNIRLNNRKIATKGWNWGEFKLENSNLCFDIDNKYAFNLPTNNINQLNVQI 154
Query 63 KNDLAIELVQDDTMDQ-SEDMLLEIRLFQPATGDDETDSHLQNLKEKLVK 111
K D+A+E D+ ++ +ED L EIR + P D+ + + QNLK L++
Sbjct 155 KTDIAMEFKNDENNNKGNEDFLAEIRFYYPHENDE--NQNFQNLKNDLLE 202
> xla:399344 ssrp1; structure specific recognition protein 1;
K09272 structure-specific recognition protein 1
Length=693
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 35/100 (35%), Positives = 55/100 (55%), Gaps = 4/100 (4%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
Y+ + +F HF ++L K++ +WG V G L+ + PAFE+ +++Q TT
Sbjct 82 YDKLFDYFKSHFSVELVEKDLCVKGWNWGSVRFGGQLLSFDIGDQPAFELPLSNVSQCTT 141
Query 61 PSKNDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDETDS 100
KN++ +E Q+ D SE L+EIR + P T DD DS
Sbjct 142 -GKNEVTLEFHQN---DDSEVSLMEIRFYVPPTQDDGGDS 177
> dre:386844 ssrp1a, ssrp1, wu:fa11b03, wu:fd10f06; structure
specific recognition protein 1a; K09272 structure-specific recognition
protein 1
Length=518
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 55/100 (55%), Gaps = 5/100 (5%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
YE I F ++K++L+ K++ +WG G+ L+ V+ P FE+ ++Q T
Sbjct 82 YEKISAFFKANYKVELEEKDMCVKGWNWGTAKFAGSLLSFDVSDSPVFEIPLSSVSQCAT 141
Query 61 PSKNDLAIELVQDDTMDQSEDMLLEIRLFQPA-TGDDETD 99
KN++ +E Q+D + S L+E+R + P TGDD +D
Sbjct 142 -GKNEVTVEFHQNDDAEVS---LMEVRFYVPPNTGDDGSD 177
> dre:449949 ssrp1b, im:7140964; structure specific recognition
protein 1b; K09272 structure-specific recognition protein
1
Length=706
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 35/111 (31%), Positives = 59/111 (53%), Gaps = 8/111 (7%)
Query 2 EDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTP 61
E I +F D++K++L K++ +WG +G L+ VN P FE+ ++Q TT
Sbjct 83 EKISEYFKDNYKVELTEKDMCVKGWNWGTAKFNGPLLSFDVNDSPTFEIPLSSVSQCTT- 141
Query 62 SKNDLAIELVQDDTMDQSEDMLLEIRLFQPAT----GDDETDSHLQNLKEK 108
KN++ +E Q+D + S L+E+R + P T G D ++ QN+ K
Sbjct 142 GKNEVTVEFHQNDDTEVS---LMEVRFYVPPTTGDEGSDPVEAFAQNVLSK 189
> hsa:6749 SSRP1, FACT, FACT80, T160; structure specific recognition
protein 1; K09272 structure-specific recognition protein
1
Length=709
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 4/99 (4%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
+E + F H++L+L K++ +WG V G L+ + P FE+ +++Q TT
Sbjct 82 FEKLSDFFKTHYRLELMEKDLCVKGWNWGTVKFGGQLLSFDIGDQPVFEIPLSNVSQCTT 141
Query 61 PSKNDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDETD 99
KN++ +E Q+D + S L+E+R + P T +D D
Sbjct 142 -GKNEVTLEFHQNDDAEVS---LMEVRFYVPPTQEDGVD 176
> mmu:20833 Ssrp1, C81323, Hmg1-rs1, Hmgi-rs3, Hmgox, T160; structure
specific recognition protein 1; K09272 structure-specific
recognition protein 1
Length=708
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 30/99 (30%), Positives = 53/99 (53%), Gaps = 4/99 (4%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
+E + F H++L+L K++ +WG V G L+ + P FE+ +++Q TT
Sbjct 82 FEKLSDFFKTHYRLELMEKDLCVKGWNWGTVKFGGQLLSFDIGDQPVFEIPLSNVSQCTT 141
Query 61 PSKNDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDETD 99
KN++ +E Q+D + S L+E+R + P T +D D
Sbjct 142 -GKNEVTLEFHQNDDAEVS---LMEVRFYVPPTQEDGVD 176
> cpv:cgd7_5280 structure-specific recognition protein 1 (SSRP1)
(recombination signal sequence recognition protein) ; K09272
structure-specific recognition protein 1
Length=523
Score = 55.1 bits (131), Expect = 5e-08, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 59/114 (51%), Gaps = 3/114 (2%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
Y I+ HF ++ + L+TK + +WGD+++ + + + G V + +I Q+
Sbjct 87 YSVIKSHFETYYGINLETKELNTKGINWGDLTIHNDTICIGNEGKVMMYVPSININQIAM 146
Query 61 PSKNDLAIELVQDDTMDQSEDMLLEIRLFQP---ATGDDETDSHLQNLKEKLVK 111
PSK++L +E + + D L+EIRLF P + D + S + L+ L+K
Sbjct 147 PSKSELVLEFNEGVNAGEDCDELMEIRLFVPNQENSLDGNSLSSAEKLRSDLLK 200
> xla:779189 raph1, MGC114656; Ras association (RalGDS/AF-6) and
pleckstrin homology domains 1
Length=365
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 30/91 (32%), Positives = 49/91 (53%), Gaps = 4/91 (4%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
Y+ + +F HF ++L K++ +WG V G L+ + PAFE+ +++Q TT
Sbjct 82 YDKLFDYFKSHFSVELVEKDLCVKGWNWGSVRFGGQLLSFDIGDQPAFELPLSNVSQCTT 141
Query 61 PSKNDLAIELVQDDTMDQSEDMLLEIRLFQP 91
KN++ +E Q D SE L+EIR + P
Sbjct 142 -GKNEVTLEFHQ---TDDSEVSLMEIRFYVP 168
> sce:YML069W POB3; Pob3p; K09272 structure-specific recognition
protein 1
Length=552
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 51/108 (47%), Gaps = 1/108 (0%)
Query 1 YEDIRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTT 60
Y I+ F F +Q++ + + +WG + N + ++NG P FE+ I
Sbjct 86 YNLIKNDFHRRFNIQVEQREHSLRGWNWGKTDLARNEMVFALNGKPTFEIPYARINNTNL 145
Query 61 PSKNDLAIEL-VQDDTMDQSEDMLLEIRLFQPATGDDETDSHLQNLKE 107
SKN++ IE +QD+ + D L+E+R + P D ++ +E
Sbjct 146 TSKNEVGIEFNIQDEEYQPAGDELVEMRFYIPGVIQTNVDENMTKKEE 193
> ath:AT3G28730 ATHMG; ATHMG (ARABIDOPSIS THALIANA HIGH MOBILITY
GROUP); transcription factor; K09272 structure-specific recognition
protein 1
Length=646
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 40/74 (54%), Gaps = 1/74 (1%)
Query 19 KNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPSKNDLAIELVQDDTMDQ 78
K ++ R+WG+V + GN L V AFEV+ D++Q KND+ +E DDT
Sbjct 100 KQLSVSGRNWGEVDLHGNTLTFLVGSKQAFEVSLADVSQTQLQGKNDVTLEFHVDDTAGA 159
Query 79 SE-DMLLEIRLFQP 91
+E D L+EI P
Sbjct 160 NEKDSLMEISFHIP 173
> cel:C32F10.5 hmg-3; HMG family member (hmg-3); K09272 structure-specific
recognition protein 1
Length=689
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 23/97 (23%), Positives = 47/97 (48%), Gaps = 4/97 (4%)
Query 4 IRRHFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQVTTPSK 63
I+ S ++ + N+ ++G + G N+ S P FE+ +++ V +K
Sbjct 84 IQSFTSSNWSKSINQSNLFINGWNYGQADVKGKNIEFSWENEPIFEIPCTNVSNVIA-NK 142
Query 64 NDLAIELVQDDTMDQSEDMLLEIRLFQPATGDDETDS 100
N+ +E Q+ +QS+ L+E+R P ++E D+
Sbjct 143 NEAILEFHQN---EQSKVQLMEMRFHMPVDLENEEDT 176
> hsa:23379 DKFZp781J2344; KIAA0947
Length=2266
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 21/36 (58%), Gaps = 1/36 (2%)
Query 7 HFSDHFKLQLQTKNIAHLSRHWGDVSMDGNNLNVSV 42
H S+H KLQ +T N HL + S+ GNNL S+
Sbjct 669 HHSEH-KLQTKTLNTLHLQSEPPECSIGGNNLENSL 703
> dre:571488 tripartite motif protein TRIM29-like
Length=542
Score = 28.5 bits (62), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 16 LQTKNIAHLSRHWGDVSMDGNNLNVSVNGCPAFEVNAQDIAQV 58
++TK+ + + VS+ G++ NV+V+ CP+F + I+Q+
Sbjct 265 VETKDHINFLKSCPSVSVSGSSDNVTVSSCPSFNEVMKSISQL 307
Lambda K H
0.315 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2062416360
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40