bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5845_orf2
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
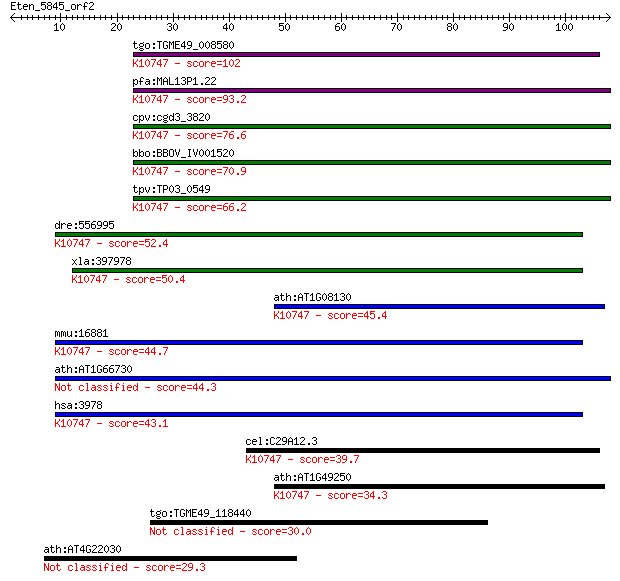
tgo:TGME49_008580 DNA ligase 1 precursor, putative (EC:3.2.1.1... 102 2e-22
pfa:MAL13P1.22 PfLigI; DNA ligase I (EC:6.5.1.1); K10747 DNA l... 93.2 2e-19
cpv:cgd3_3820 DNA LIGASE I ; K10747 DNA ligase 1 [EC:6.5.1.1] 76.6 2e-14
bbo:BBOV_IV001520 21.m02964; DNA ligase I (EC:6.5.1.1); K10747... 70.9 9e-13
tpv:TP03_0549 DNA ligase I (EC:6.5.1.1); K10747 DNA ligase 1 [... 66.2 2e-11
dre:556995 lig1, wu:fc54a11; si:dkeyp-35b8.5 (EC:6.5.1.1); K10... 52.4 3e-07
xla:397978 lig1, ligI; ligase I, DNA, ATP-dependent (EC:6.5.1.... 50.4 1e-06
ath:AT1G08130 ATLIG1; ATLIG1 (ARABIDOPSIS THALIANA DNA LIGASE ... 45.4 4e-05
mmu:16881 Lig1, AL033288, LigI; ligase I, DNA, ATP-dependent (... 44.7 7e-05
ath:AT1G66730 ATP dependent DNA ligase family protein 44.3 9e-05
hsa:3978 LIG1, MGC117397, MGC130025; ligase I, DNA, ATP-depend... 43.1 2e-04
cel:C29A12.3 lig-1; LIGase family member (lig-1); K10747 DNA l... 39.7 0.002
ath:AT1G49250 ATP dependent DNA ligase family protein; K10747 ... 34.3 0.089
tgo:TGME49_118440 DEAH-box RNA/DNA helicase, putative (EC:3.4.... 30.0 1.7
ath:AT4G22030 F-box family protein 29.3 3.5
> tgo:TGME49_008580 DNA ligase 1 precursor, putative (EC:3.2.1.11
6.5.1.1); K10747 DNA ligase 1 [EC:6.5.1.1]
Length=1112
Score = 102 bits (255), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 43/83 (51%), Positives = 62/83 (74%), Gaps = 0/83 (0%)
Query 23 FSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAPDFL 82
F L A +IE +K SG+GS+KKQT++L N FR+L+FY P+ L A+Y CLN+ APD+L
Sbjct 415 FEALVNAFDKIEQMKASGTGSNKKQTVVLTNLFRLLLFYAPKTLHKAIYICLNKVAPDYL 474
Query 83 DAEAGVGDSILFKAVAETYGVSD 105
E GVG+S++ +A+AETYG ++
Sbjct 475 GQEVGVGESLILRAMAETYGRTE 497
> pfa:MAL13P1.22 PfLigI; DNA ligase I (EC:6.5.1.1); K10747 DNA
ligase 1 [EC:6.5.1.1]
Length=912
Score = 93.2 bits (230), Expect = 2e-19, Method: Composition-based stats.
Identities = 40/85 (47%), Positives = 61/85 (71%), Gaps = 0/85 (0%)
Query 23 FSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAPDFL 82
F+ LT A +IE LK SG+GS K ++IL+N FR+L++Y P L AVY LN+ APD+L
Sbjct 177 FTFLTNAFNQIEELKGSGTGSKKNVSIILSNIFRVLIYYSPNDLIPAVYITLNKVAPDYL 236
Query 83 DAEAGVGDSILFKAVAETYGVSDNS 107
+ EAGVG++++ K ++E Y +++S
Sbjct 237 NVEAGVGEALILKTMSEAYSRTESS 261
> cpv:cgd3_3820 DNA LIGASE I ; K10747 DNA ligase 1 [EC:6.5.1.1]
Length=825
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 46/85 (54%), Positives = 59/85 (69%), Gaps = 3/85 (3%)
Query 23 FSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAPDFL 82
FSVLT+A RIE LK SGSGS K +ILAN FR+++ + P L AVY C+N+ APD+
Sbjct 166 FSVLTDAFSRIEQLKGSGSGSKKGCIVILANLFRLIIHHNPSDLIDAVYICMNKVAPDYE 225
Query 83 DAEAGVGDSILFKAVAETYGVSDNS 107
E+GVGDSIL K ++E+ SD S
Sbjct 226 GKESGVGDSILIKCISES---SDKS 247
> bbo:BBOV_IV001520 21.m02964; DNA ligase I (EC:6.5.1.1); K10747
DNA ligase 1 [EC:6.5.1.1]
Length=773
Score = 70.9 bits (172), Expect = 9e-13, Method: Composition-based stats.
Identities = 33/85 (38%), Positives = 55/85 (64%), Gaps = 0/85 (0%)
Query 23 FSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAPDFL 82
F+ L E L ++E SG+GS K +L+N FR+L+++ P + A+Y LNR + ++
Sbjct 150 FNFLAEILQKVEDAFGSGTGSRKYVFTLLSNFFRVLIYHQPSDVIPAMYLMLNRISAEYE 209
Query 83 DAEAGVGDSILFKAVAETYGVSDNS 107
E GVGD++L KA+A++YG S+ +
Sbjct 210 GEEFGVGDTMLIKAMAQSYGKSEKA 234
> tpv:TP03_0549 DNA ligase I (EC:6.5.1.1); K10747 DNA ligase 1
[EC:6.5.1.1]
Length=858
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 29/85 (34%), Positives = 52/85 (61%), Gaps = 0/85 (0%)
Query 23 FSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAPDFL 82
FS+++ L + + + +G GS K +LAN +R+L++Y P + Y LNR P++
Sbjct 215 FSLVSSVLQKSDDIFCTGKGSRKSVLTLLANFYRVLIYYNPPDVIPVTYLLLNRICPEYE 274
Query 83 DAEAGVGDSILFKAVAETYGVSDNS 107
+ E GVGDS++ K+++E Y S+ +
Sbjct 275 NIEIGVGDSLIIKSMSEAYSKSEKN 299
> dre:556995 lig1, wu:fc54a11; si:dkeyp-35b8.5 (EC:6.5.1.1); K10747
DNA ligase 1 [EC:6.5.1.1]
Length=1058
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 28/94 (29%), Positives = 47/94 (50%), Gaps = 12/94 (12%)
Query 9 KQRQRGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAA 68
K+ Q+ L + R+F + E GR++ ++ L+N R ++ P L
Sbjct 424 KKGQKVPYLAVARTFEKIEEDSGRLKNIET------------LSNFLRSVILLSPDDLLC 471
Query 69 AVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
VY CLN+ P + E G+G+++L KAVA+ G
Sbjct 472 CVYLCLNQLGPAYQGLELGIGETVLMKAVAQATG 505
> xla:397978 lig1, ligI; ligase I, DNA, ATP-dependent (EC:6.5.1.1);
K10747 DNA ligase 1 [EC:6.5.1.1]
Length=1070
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 27/91 (29%), Positives = 45/91 (49%), Gaps = 12/91 (13%)
Query 12 QRGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAAAVY 71
Q+ L + R+F + E R++ ++ L+N R ++ P+ L +Y
Sbjct 438 QKVPYLAVARTFERIEEESARLKNVET------------LSNFLRSVIALTPEDLLPCIY 485
Query 72 FCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
CLNR P + E G+G++IL KAVA+ G
Sbjct 486 LCLNRLGPAYEGLELGIGETILMKAVAQATG 516
> ath:AT1G08130 ATLIG1; ATLIG1 (ARABIDOPSIS THALIANA DNA LIGASE
1); ATP binding / DNA binding / DNA ligase (ATP); K10747 DNA
ligase 1 [EC:6.5.1.1]
Length=790
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 33/59 (55%), Gaps = 0/59 (0%)
Query 48 TLILANAFRILVFYCPQALAAAVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYGVSDN 106
T IL N R ++ P+ L A VY N AP E G+G+S + KA++E +G +++
Sbjct 197 TDILCNMLRTVIATTPEDLVATVYLSANEIAPAHEGVELGIGESTIIKAISEAFGRTED 255
> mmu:16881 Lig1, AL033288, LigI; ligase I, DNA, ATP-dependent
(EC:6.5.1.1); K10747 DNA ligase 1 [EC:6.5.1.1]
Length=932
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 40/94 (42%), Gaps = 12/94 (12%)
Query 9 KQRQRGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAA 68
K Q+ L + R+F + E R+ K L+N R +V P L
Sbjct 296 KHGQKVPFLAVARTFEKIEEVSARL------------KMVETLSNLLRSVVALSPPDLLP 343
Query 69 AVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
+Y LNR P E GVGD +L KAVA+ G
Sbjct 344 VLYLSLNRLGPPQQGLELGVGDGVLLKAVAQATG 377
> ath:AT1G66730 ATP dependent DNA ligase family protein
Length=1417
Score = 44.3 bits (103), Expect = 9e-05, Method: Composition-based stats.
Identities = 26/99 (26%), Positives = 47/99 (47%), Gaps = 12/99 (12%)
Query 9 KQRQRGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAA 68
++ Q + L R+F+ + G+I+ + +L N FR L P+ +
Sbjct 760 REGQPAPYIHLVRTFASVESEKGKIKAMS------------MLCNMFRSLFALSPEDVLP 807
Query 69 AVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYGVSDNS 107
AVY C N+ A D + E +G S++ A+ E G+S ++
Sbjct 808 AVYLCTNKIAADHENIELNIGGSLISSALEEACGISRST 846
> hsa:3978 LIG1, MGC117397, MGC130025; ligase I, DNA, ATP-dependent
(EC:6.5.1.1); K10747 DNA ligase 1 [EC:6.5.1.1]
Length=919
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 40/94 (42%), Gaps = 12/94 (12%)
Query 9 KQRQRGQRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLILANAFRILVFYCPQALAA 68
K Q+ L + R+F + E R+ ++ L+N R +V P L
Sbjct 282 KPGQKVPYLAVARTFEKIEEVSARLRMVET------------LSNLLRSVVALSPPDLLP 329
Query 69 AVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
+Y LN P E GVGD +L KAVA+ G
Sbjct 330 VLYLSLNHLGPPQQGLELGVGDGVLLKAVAQATG 363
> cel:C29A12.3 lig-1; LIGase family member (lig-1); K10747 DNA
ligase 1 [EC:6.5.1.1]
Length=773
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 43 SSKKQTLILANAFRILVFYCPQALAAAVYFCLNRTAPDFLDAEAGVGDSILFKAVAETYG 102
S KK+ LA F ++ + P L A VY +N+ P + E GV ++ L KAVA+ G
Sbjct 35 SGKKKVDELAQFFTKVLDFSPDDLTACVYMSVNQLGPSYEGLELGVAENSLIKAVAKATG 94
Query 103 VSD 105
++
Sbjct 95 RTE 97
> ath:AT1G49250 ATP dependent DNA ligase family protein; K10747
DNA ligase 1 [EC:6.5.1.1]
Length=657
Score = 34.3 bits (77), Expect = 0.089, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 48 TLILANAFRILVFYCPQALAAAVYFCLNRTAPDFLDAEAGVGD-SILFKAVAETYGVSDN 106
T IL N R ++ P L VY N AP + G+G S + KA++E +G +++
Sbjct 65 THILCNMLRTVIATTPDDLLPTVYLAANEIAPAHEGIKLGMGKGSYIIKAISEAFGRTES 124
> tgo:TGME49_118440 DEAH-box RNA/DNA helicase, putative (EC:3.4.22.44
3.6.1.15)
Length=2234
Score = 30.0 bits (66), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 29/63 (46%), Gaps = 3/63 (4%)
Query 26 LTEALGRIEGLKNSGSGSSKKQT---LILANAFRILVFYCPQALAAAVYFCLNRTAPDFL 82
L E LGR G + SG + K+T + F ++ P ALAA + L+ L
Sbjct 367 LEEPLGRSVGYRISGDAVTSKETKVCFVTTGYFLQVLINQPHALAALTHIILDEVHERDL 426
Query 83 DAE 85
DA+
Sbjct 427 DAD 429
> ath:AT4G22030 F-box family protein
Length=626
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 27/48 (56%), Gaps = 3/48 (6%)
Query 7 RKKQRQRG---QRLGLGRSFSVLTEALGRIEGLKNSGSGSSKKQTLIL 51
R++Q++RG Q L R +VL E + R+E KN G + +L+L
Sbjct 70 RQRQKKRGCDDQVLQRSRLMAVLEEVIDRVEMHKNIGEQRNNWNSLLL 117
Lambda K H
0.322 0.137 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40