bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5840_orf4
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
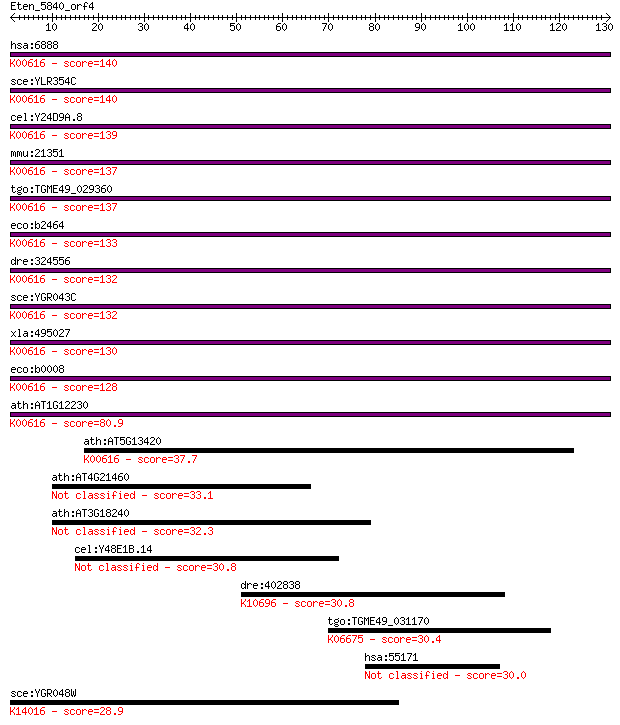
hsa:6888 TALDO1, TAL, TAL-H, TALDOR, TALH; transaldolase 1 (EC... 140 7e-34
sce:YLR354C TAL1; Tal1p (EC:2.2.1.2); K00616 transaldolase [EC... 140 1e-33
cel:Y24D9A.8 hypothetical protein; K00616 transaldolase [EC:2.... 139 3e-33
mmu:21351 Taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transal... 137 6e-33
tgo:TGME49_029360 transaldolase, putative (EC:2.2.1.2); K00616... 137 1e-32
eco:b2464 talA, ECK2459, JW2448; transaldolase A (EC:2.2.1.2);... 133 1e-31
dre:324556 taldo1, MGC56232, Tal, wu:fc32g02, zgc:56232, zgc:6... 132 3e-31
sce:YGR043C NQM1; Nqm1p (EC:2.2.1.2); K00616 transaldolase [EC... 132 3e-31
xla:495027 taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transa... 130 8e-31
eco:b0008 talB, ECK0008, JW0007, yaaK; transaldolase B (EC:2.2... 128 5e-30
ath:AT1G12230 transaldolase, putative; K00616 transaldolase [E... 80.9 1e-15
ath:AT5G13420 transaldolase, putative; K00616 transaldolase [E... 37.7 0.009
ath:AT4G21460 hypothetical protein 33.1 0.25
ath:AT3G18240 hypothetical protein 32.3 0.37
cel:Y48E1B.14 hypothetical protein 30.8 1.1
dre:402838 hypothetical protein LOC402838; K10696 E3 ubiquitin... 30.8 1.2
tgo:TGME49_031170 chromosome condensation protein, putative ; ... 30.4 1.5
hsa:55171 TBCCD1, FLJ10560; TBCC domain containing 1 30.0
sce:YGR048W UFD1; Protein that interacts with Cdc48p and Npl4p... 28.9 3.7
> hsa:6888 TALDO1, TAL, TAL-H, TALDOR, TALH; transaldolase 1 (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=337
Score = 140 bits (354), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 70/130 (53%), Positives = 92/130 (70%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF AI E+ P DATTNP+L+L A Y+ L ++AI ++ G E ++ A+
Sbjct 27 DTGDFHAIDEYKPQDATTNPSLILAAAQMPAYQELVEEAIAYGRKL--GGSQEDQIKNAI 84
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
D++ V FG E+L+ IPG VSTE+ A LSFD + VARARR+IELYK GISKDRILIK++
Sbjct 85 DKLFVLFGAEILKKIPGRVSTEVDARLSFDKDAMVARARRLIELYKEAGISKDRILIKLS 144
Query 121 ATWEGIQAAK 130
+TWEGIQA K
Sbjct 145 STWEGIQAGK 154
> sce:YLR354C TAL1; Tal1p (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=335
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 67/130 (51%), Positives = 94/130 (72%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF +I +F P D+TTNP+L+L A Y L A+ ++ GK E VE AV
Sbjct 29 DTGDFGSIAKFQPQDSTTNPSLILAAAKQPTYAKLIDVAVEYGKK--HGKTTEEQVENAV 86
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
DR+LV FG E+L+I+PG VSTE+ A LSFDT+ ++ +AR II+L++ +G+SK+R+LIK+A
Sbjct 87 DRLLVEFGKEILKIVPGRVSTEVDARLSFDTQATIEKARHIIKLFEQEGVSKERVLIKIA 146
Query 121 ATWEGIQAAK 130
+TWEGIQAAK
Sbjct 147 STWEGIQAAK 156
> cel:Y24D9A.8 hypothetical protein; K00616 transaldolase [EC:2.2.1.2]
Length=319
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 69/130 (53%), Positives = 94/130 (72%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF AIKEF PTDATTNP+L+L A +Y L +++ A+E + G + ++ A+
Sbjct 16 DTGDFNAIKEFQPTDATTNPSLILAASKMEQYAALIDQSVAYAKEHASGH--QEVLQAAM 73
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
DR+ V FG E+L+ IPG VSTE+ A LSFDT+ S+ RA +I Y+ +GISKDRILIK+A
Sbjct 74 DRLFVVFGKEILKTIPGRVSTEVDARLSFDTQASIDRALGLIAQYEKEGISKDRILIKLA 133
Query 121 ATWEGIQAAK 130
+TWEGI+AAK
Sbjct 134 STWEGIRAAK 143
> mmu:21351 Taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transaldolase
[EC:2.2.1.2]
Length=337
Score = 137 bits (346), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 68/130 (52%), Positives = 91/130 (70%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF AI E+ P DATTNP+L+L A Y+ L ++AI ++ G E ++ A+
Sbjct 27 DTGDFNAIDEYKPQDATTNPSLILAAAQMPAYQELVEEAIAYGKKL--GGPQEEQIKNAI 84
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
D++ V FG E+L+ IPG VSTE+ A LSFD + VARARR+IELYK G+ KDRILIK++
Sbjct 85 DKLFVLFGAEILKKIPGRVSTEVDARLSFDKDAMVARARRLIELYKEAGVGKDRILIKLS 144
Query 121 ATWEGIQAAK 130
+TWEGIQA K
Sbjct 145 STWEGIQAGK 154
> tgo:TGME49_029360 transaldolase, putative (EC:2.2.1.2); K00616
transaldolase [EC:2.2.1.2]
Length=364
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 65/131 (49%), Positives = 97/131 (74%), Gaps = 1/131 (0%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEAS-KGKDLEATVEEA 59
D+ DF A+K++ P DATTNP+L+L+A S +Y +L +A+ A++ K + + VEE
Sbjct 49 DTGDFVALKKYVPHDATTNPSLLLKAASMPQYAHLMDEAVEVAKKVHGKVEPDDELVEEV 108
Query 60 VDRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKV 119
+D++ V FG+E+L+I+PG+VSTE+ A LSFD + SV +AR++I +YK GI KDR+LIK+
Sbjct 109 IDQLFVRFGVEILKIVPGVVSTEVSAALSFDVQASVEKARKLILMYKEHGIEKDRVLIKL 168
Query 120 AATWEGIQAAK 130
A TWEG +AAK
Sbjct 169 ATTWEGCEAAK 179
> eco:b2464 talA, ECK2459, JW2448; transaldolase A (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=316
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 65/130 (50%), Positives = 92/130 (70%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
DS D E+I+ + P DATTNP+L+L+A S+Y++L AI A GK E V A
Sbjct 16 DSGDIESIRHYHPQDATTNPSLLLKAAGLSQYEHLIDDAI--AWGKKNGKTQEQQVVAAC 73
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
D++ V FG E+L+I+PG VSTE+ A LSFD E S+ +AR +++LY+ +G+ K RILIK+A
Sbjct 74 DKLAVNFGAEILKIVPGRVSTEVDARLSFDKEKSIEKARHLVDLYQQQGVEKSRILIKLA 133
Query 121 ATWEGIQAAK 130
+TWEGI+AA+
Sbjct 134 STWEGIRAAE 143
> dre:324556 taldo1, MGC56232, Tal, wu:fc32g02, zgc:56232, zgc:66054;
transaldolase 1 (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=337
Score = 132 bits (332), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 64/130 (49%), Positives = 90/130 (69%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF AI+E+ P DATTNP+L+L A Y+ L +AI + G + V A+
Sbjct 27 DTGDFNAIEEYKPQDATTNPSLILAAAKMPAYQPLVDQAIKYG--TANGGTEDEQVTNAM 84
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
D++ V FG+E+L+ +PG VSTE+ A LSFD + V+RARR+I LY+ GISK+RILIK++
Sbjct 85 DKLFVNFGLEILKKVPGRVSTEVDARLSFDKDAMVSRARRLISLYEDAGISKERILIKLS 144
Query 121 ATWEGIQAAK 130
+TWEGIQA +
Sbjct 145 STWEGIQAGR 154
> sce:YGR043C NQM1; Nqm1p (EC:2.2.1.2); K00616 transaldolase [EC:2.2.1.2]
Length=333
Score = 132 bits (332), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 62/130 (47%), Positives = 90/130 (69%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
DS DFEAI ++ P D+TTNP+L+L A KY A+ ++ GK +E A+
Sbjct 29 DSGDFEAISKYEPQDSTTNPSLILAASKLEKYARFIDAAVEYGRK--HGKTDHEKIENAM 86
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
D++LV FG ++L+++PG VSTE+ A LSFD + +V +A II+LYK G+ K+R+LIK+A
Sbjct 87 DKILVEFGTQILKVVPGRVSTEVDARLSFDKKATVKKALHIIKLYKDAGVPKERVLIKIA 146
Query 121 ATWEGIQAAK 130
+TWEGIQAA+
Sbjct 147 STWEGIQAAR 156
> xla:495027 taldo1; transaldolase 1 (EC:2.2.1.2); K00616 transaldolase
[EC:2.2.1.2]
Length=338
Score = 130 bits (328), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 68/134 (50%), Positives = 89/134 (66%), Gaps = 10/134 (7%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ DF AI+E+ P DATTNP+L+L A Y+ L AI GK+L + EE +
Sbjct 28 DTGDFNAIEEYKPQDATTNPSLILAAAQMPDYQGLVNDAI------QYGKNLGGSEEEQI 81
Query 61 ----DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRIL 116
D++ V FG+E+L+ IPG VSTE+ A LSFD +G V RA+R+I LYK GI K RIL
Sbjct 82 NNIMDKLFVLFGVEILKKIPGRVSTEVDARLSFDKDGMVERAKRLIALYKEAGIDKKRIL 141
Query 117 IKVAATWEGIQAAK 130
IK+++TWEGIQA K
Sbjct 142 IKLSSTWEGIQAGK 155
> eco:b0008 talB, ECK0008, JW0007, yaaK; transaldolase B (EC:2.2.1.2);
K00616 transaldolase [EC:2.2.1.2]
Length=317
Score = 128 bits (321), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 60/130 (46%), Positives = 92/130 (70%), Gaps = 2/130 (1%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D+ D A+K + P DATTNP+L+L A +Y+ L A+ A++ S D + +A
Sbjct 17 DTGDIAAMKLYQPQDATTNPSLILNAAQIPEYRKLIDDAVAWAKQQSN--DRAQQIVDAT 74
Query 61 DRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVA 120
D++ V G+E+L+++PG +STE+ A LS+DTE S+A+A+R+I+LY GIS DRILIK+A
Sbjct 75 DKLAVNIGLEILKLVPGRISTEVDARLSYDTEASIAKAKRLIKLYNDAGISNDRILIKLA 134
Query 121 ATWEGIQAAK 130
+TW+GI+AA+
Sbjct 135 STWQGIRAAE 144
> ath:AT1G12230 transaldolase, putative; K00616 transaldolase
[EC:2.2.1.2]
Length=405
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 43/133 (32%), Positives = 74/133 (55%), Gaps = 5/133 (3%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEAT---VE 57
D+ F+ + F PT AT + L+L +F+ A++ A S LE T +
Sbjct 83 DTVVFDDFERFPPTAATVSSALLLGICGLPD--TIFRNAVDMALADSSCAGLETTESRLS 140
Query 58 EAVDRVLVGFGMELLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILI 117
++ +V G +L++++PG VSTE+ A L++DT G + + ++ LY + DR+L
Sbjct 141 CFFNKAIVNVGGDLVKLVPGRVSTEVDARLAYDTNGIIRKVHDLLRLYNEIDVPHDRLLF 200
Query 118 KVAATWEGIQAAK 130
K+ ATW+GI+AA+
Sbjct 201 KIPATWQGIEAAR 213
> ath:AT5G13420 transaldolase, putative; K00616 transaldolase
[EC:2.2.1.2]
Length=438
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 56/114 (49%), Gaps = 20/114 (17%)
Query 17 TTNPTLVLQAVSNSK-YKNLFQKAINEAQEASKGKDLEATVEEAVDRVLVGFGMELLEII 75
T+NP + +A+S S Y + F+ + GKD+E+ E V + + +L E I
Sbjct 112 TSNPAIFQKAISTSNAYNDQFRTLVES------GKDIESAYWELVVKDIQD-ACKLFEPI 164
Query 76 -------PGLVSTEIPAELSFDTEGSVARARRIIELYKSKGISKDRILIKVAAT 122
G VS E+ L+ DT+G+V A+ Y SK +++ + IK+ AT
Sbjct 165 YDQTEGADGYVSVEVSPRLADDTQGTVEAAK-----YLSKVVNRRNVYIKIPAT 213
> ath:AT4G21460 hypothetical protein
Length=415
Score = 33.1 bits (74), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 30/56 (53%), Gaps = 4/56 (7%)
Query 10 EFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAVDRVLV 65
E SPTD++ LV++ VSN + K+ K+ NE E + L +E + R LV
Sbjct 56 ESSPTDSSDKKDLVVEDVSNKELKSRIDKSFNEGNEDA----LPGVIEALLQRRLV 107
> ath:AT3G18240 hypothetical protein
Length=419
Score = 32.3 bits (72), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 35/73 (47%), Gaps = 8/73 (10%)
Query 10 EFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAVDRVLVGFGM 69
E SPTD+ LV++ VSN + K+ +K NE E + L +E + R LV
Sbjct 60 ESSPTDSPEKKDLVVEDVSNKELKSRIEKYFNEGNEDA----LPGVIEALLQRRLVDKHA 115
Query 70 ----ELLEIIPGL 78
ELLE I L
Sbjct 116 ETDDELLEKIESL 128
> cel:Y48E1B.14 hypothetical protein
Length=971
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 0/57 (0%)
Query 15 DATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAVDRVLVGFGMEL 71
DATT+ +L AV +S Y N F E+ + +D+ A D +L+ F L
Sbjct 771 DATTSYSLATNAVYSSVYGNNFDGVFKESCRSLGDEDMRLWTRSAYDSILLLFWQPL 827
> dre:402838 hypothetical protein LOC402838; K10696 E3 ubiquitin-protein
ligase BRE1 [EC:6.3.2.19]
Length=1013
Score = 30.8 bits (68), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query 51 DLEATVEEAVDRVLVGFGME-LLEIIPGLVSTEIPAELSFDTEGSVARARRIIELYKS 107
D ++ E A DR G G+ L + S EI AEL E S +A R++E+Y++
Sbjct 148 DGDSNQERAKDRGQQGEGVSSFLATLASSTSEEIDAELQERVESSCKQASRVVEIYEN 205
> tgo:TGME49_031170 chromosome condensation protein, putative
; K06675 structural maintenance of chromosome 4
Length=1640
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 33/57 (57%), Gaps = 10/57 (17%)
Query 70 ELLEIIPG---LVSTEIPAELSFDTE----GSVARARRIIELYKSKG--ISKDRILI 117
E E+IPG +VS E+ + S TE G A+ R+++EL KSKG + +R LI
Sbjct 247 ETYEVIPGSQFVVSREV-SRASNSTEYRVNGQRAQQRQVVELLKSKGLDLQNNRFLI 302
> hsa:55171 TBCCD1, FLJ10560; TBCC domain containing 1
Length=557
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 78 LVSTEIPAELSFDTEGSVARARRIIELYK 106
LVS E LSF EG+++RAR+I L++
Sbjct 203 LVSREAVVALSFLIEGTISRARKIYPLHE 231
> sce:YGR048W UFD1; Protein that interacts with Cdc48p and Npl4p,
involved in recognition of polyubiquitinated proteins and
their presentation to the 26S proteasome for degradation;
involved in transporting proteins from the ER to the cytosol;
K14016 ubiquitin fusion degradation protein 1
Length=361
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 42/89 (47%), Gaps = 13/89 (14%)
Query 1 DSADFEAIKEFSPTDATTNPTLVLQAVSNSKYKNLFQKAINEAQEASKGKDLEATVEEAV 60
D A+F K F P A + +++ N +Y LF+ NE + G LE EE
Sbjct 40 DDANFGG-KIFLPPSALSKLSML-----NIRYPMLFKLTANETGRVTHGGVLEFIAEEG- 92
Query 61 DRV-LVGFGMELLEIIPG----LVSTEIP 84
RV L + ME L I PG + ST++P
Sbjct 93 -RVYLPQWMMETLGIQPGSLLQISSTDVP 120
Lambda K H
0.312 0.129 0.340
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40