bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5830_orf1
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
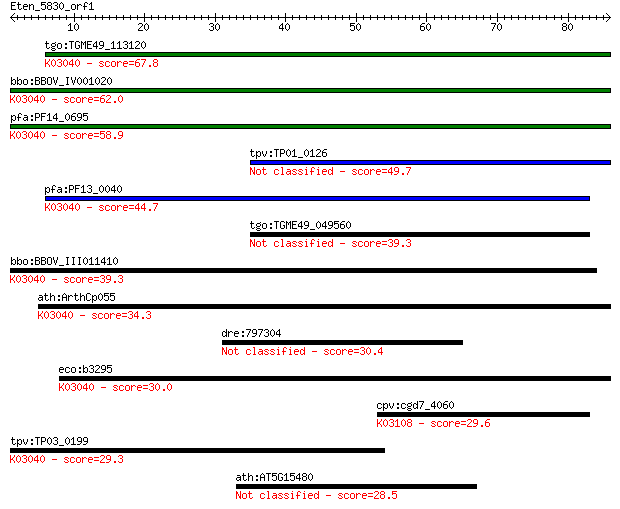
tgo:TGME49_113120 RNA polymerase Rpb3/Rpb11 dimerization domai... 67.8 8e-12
bbo:BBOV_IV001020 21.m03001; DNA-directed RNA polymerase, alph... 62.0 5e-10
pfa:PF14_0695 DNA-directed RNA polymerase, alpha subunit, puta... 58.9 3e-09
tpv:TP01_0126 DNA-directed RNA polymerase, subunit alpha 49.7 2e-06
pfa:PF13_0040 DNA-directed RNA polymerase alpha chain, putativ... 44.7 8e-05
tgo:TGME49_049560 DNA-directed RNA polymerase alpha chain, put... 39.3 0.003
bbo:BBOV_III011410 17.m07973; hypothetical protein; K03040 DNA... 39.3 0.003
ath:ArthCp055 rpoA; RNA polymerase alpha chain; K03040 DNA-dir... 34.3 0.10
dre:797304 smc6; structural maintenance of chromosomes 6 30.4
eco:b3295 rpoA, ECK3282, JW3257, pez, phs, sez; RNA polymerase... 30.0 2.1
cpv:cgd7_4060 signal recognition particle SPR72 ; K03108 signa... 29.6 2.6
tpv:TP03_0199 DNA-directed RNA polymerase subunit alpha (EC:2.... 29.3 3.0
ath:AT5G15480 zinc finger (C2H2 type) family protein 28.5 6.0
> tgo:TGME49_113120 RNA polymerase Rpb3/Rpb11 dimerization domain-containing
protein (EC:2.7.7.6); K03040 DNA-directed RNA
polymerase subunit alpha [EC:2.7.7.6]
Length=623
Score = 67.8 bits (164), Expect = 8e-12, Method: Composition-based stats.
Identities = 32/80 (40%), Positives = 50/80 (62%), Gaps = 0/80 (0%)
Query 6 VPGMREDVLELALNLKTLSLRSGAPFARCRVRVHARGPLKLLGGHISFPSFLKCLNKENY 65
+P +RED+LE+ALNLK ++ + P VRV A GP L+ G I +PSF++ N E++
Sbjct 228 LPNVREDLLEIALNLKGVAFETLRPRESACVRVKAVGPQLLVAGMIDWPSFVRVGNPEHF 287
Query 66 ICQLAAAAAVCFEFQVEWGR 85
I ++ + E ++EWGR
Sbjct 288 IAKVEEGGVLDLEMKLEWGR 307
> bbo:BBOV_IV001020 21.m03001; DNA-directed RNA polymerase, alpha
subunit; K03040 DNA-directed RNA polymerase subunit alpha
[EC:2.7.7.6]
Length=676
Score = 62.0 bits (149), Expect = 5e-10, Method: Composition-based stats.
Identities = 27/85 (31%), Positives = 47/85 (55%), Gaps = 0/85 (0%)
Query 1 NGALSVPGMREDVLELALNLKTLSLRSGAPFARCRVRVHARGPLKLLGGHISFPSFLKCL 60
N ++ G+ +D++ +ALNL ++L + P RVR +GP ++ G I +PSF+K
Sbjct 299 NEDTTIDGVTDDLMNIALNLNEVALETLEPGGEARVRTVIKGPAQVCAGSIEWPSFVKIA 358
Query 61 NKENYICQLAAAAAVCFEFQVEWGR 85
+ YI + + E ++EWGR
Sbjct 359 TPDAYITNVEEGGTLDIEIKIEWGR 383
> pfa:PF14_0695 DNA-directed RNA polymerase, alpha subunit, putative;
K03040 DNA-directed RNA polymerase subunit alpha [EC:2.7.7.6]
Length=861
Score = 58.9 bits (141), Expect = 3e-09, Method: Composition-based stats.
Identities = 28/85 (32%), Positives = 48/85 (56%), Gaps = 0/85 (0%)
Query 1 NGALSVPGMREDVLELALNLKTLSLRSGAPFARCRVRVHARGPLKLLGGHISFPSFLKCL 60
N + ++ED+LE+ALN+ + + S +R+ +GPL L+ G I PS LK +
Sbjct 521 NEDTKINNVQEDLLEIALNISDVCIYSKEINLESNIRLIFKGPLMLVAGMIPLPSHLKII 580
Query 61 NKENYICQLAAAAAVCFEFQVEWGR 85
NKE+YIC + + ++E+G+
Sbjct 581 NKEHYICTIKEDGFIDISIKIEYGK 605
> tpv:TP01_0126 DNA-directed RNA polymerase, subunit alpha
Length=746
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 22/51 (43%), Positives = 30/51 (58%), Gaps = 0/51 (0%)
Query 35 RVRVHARGPLKLLGGHISFPSFLKCLNKENYICQLAAAAAVCFEFQVEWGR 85
RVR +GP L G + +PSF+K N E YI +L A + E ++EWGR
Sbjct 405 RVRTVIKGPAMLCAGALEWPSFVKLCNPECYITKLEEGAELDLEIKIEWGR 455
> pfa:PF13_0040 DNA-directed RNA polymerase alpha chain, putative
(EC:2.7.7.6); K03040 DNA-directed RNA polymerase subunit
alpha [EC:2.7.7.6]
Length=619
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 21/84 (25%), Positives = 48/84 (57%), Gaps = 10/84 (11%)
Query 6 VPGMREDVLELALNLKTLSLRSGAP-------FARCRVRVHARGPLKLLGGHISFPSFLK 58
+ G+RE+ +LA NL+ ++ ++ P + + R+ +GP+ ++ GH+ P ++
Sbjct 297 IVGVRENFFDLAQNLRQITFKN-VPEDVDMDNYMIGKFRI--KGPMIVVAGHMQLPKNVE 353
Query 59 CLNKENYICQLAAAAAVCFEFQVE 82
+N+ YIC +AA + + + ++E
Sbjct 354 IVNRNQYICSVAAGSYLSMDVKIE 377
> tgo:TGME49_049560 DNA-directed RNA polymerase alpha chain, putative
(EC:2.7.7.6)
Length=836
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 35 RVRVHARGPLKLLGGHISFPSFLKCLNKENYICQLAAAAAVCFEFQVE 82
R ++ RGPL + GHI P ++ +NK Y+ + A V +F++E
Sbjct 283 RAKLRVRGPLVAVAGHIQLPEGVEVVNKNQYLFTVNAGYYVNMDFKIE 330
> bbo:BBOV_III011410 17.m07973; hypothetical protein; K03040 DNA-directed
RNA polymerase subunit alpha [EC:2.7.7.6]
Length=610
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 47/104 (45%), Gaps = 21/104 (20%)
Query 1 NGALSVPGMREDVLELALNLKTLSLRS---GAPFARCRV-RVHARGPLKLLGGHISFPSF 56
N S+ G+RED +L+ NL + R+ A F R + R+ +GP+ + GHI
Sbjct 264 NEFYSIVGLREDFFDLSRNLTKVIFRNVPMTATFHRPLIGRLRLKGPIIAVAGHIELDQT 323
Query 57 -----------------LKCLNKENYICQLAAAAAVCFEFQVEW 83
++ +NK YIC L+ A + E ++E+
Sbjct 324 FQKEGVGSSSGSKDDRRIEIVNKNQYICALSRDAYLDMEVKIEY 367
> ath:ArthCp055 rpoA; RNA polymerase alpha chain; K03040 DNA-directed
RNA polymerase subunit alpha [EC:2.7.7.6]
Length=329
Score = 34.3 bits (77), Expect = 0.10, Method: Composition-based stats.
Identities = 20/81 (24%), Positives = 38/81 (46%), Gaps = 1/81 (1%)
Query 5 SVPGMREDVLELALNLKTLSLRSGAPFARCRVRVHARGPLKLLGGHISFPSFLKCLNKEN 64
++ G++E V E+ +NL + LRS R + +GP + I P ++ ++
Sbjct 76 NIAGIQESVHEILMNLNEIVLRSNLYGTR-NALICVQGPGYITARDIILPPAVEIIDNTQ 134
Query 65 YICQLAAAAAVCFEFQVEWGR 85
+I L +C E ++E R
Sbjct 135 HIATLTEPIDLCIELKIERNR 155
> dre:797304 smc6; structural maintenance of chromosomes 6
Length=948
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 31 FARCRVRVHARGPLKLLGGHISFPSFLKCLNKEN 64
FA+ R + GP+ H FPS L CLN EN
Sbjct 432 FAQGRFKKKPVGPIGRGVHHPEFPSVLDCLNIEN 465
> eco:b3295 rpoA, ECK3282, JW3257, pez, phs, sez; RNA polymerase,
alpha subunit (EC:2.7.7.6); K03040 DNA-directed RNA polymerase
subunit alpha [EC:2.7.7.6]
Length=329
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 22/80 (27%), Positives = 40/80 (50%), Gaps = 4/80 (5%)
Query 8 GMREDVLELALNLKTLSLR-SGAPFARCRVRVHARGPLKLLGGHISFPSFLKCLNKENYI 66
G++ED+LE+ LNLK L++R G + GP+ I+ ++ + ++ I
Sbjct 73 GVQEDILEILLNLKGLAVRVQGKDEVILTLNKSGIGPVT--AADITHDGDVEIVKPQHVI 130
Query 67 CQLA-AAAAVCFEFQVEWGR 85
C L A++ +V+ GR
Sbjct 131 CHLTDENASISMRIKVQRGR 150
> cpv:cgd7_4060 signal recognition particle SPR72 ; K03108 signal
recognition particle subunit SRP72
Length=680
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 16/30 (53%), Gaps = 0/30 (0%)
Query 53 FPSFLKCLNKENYICQLAAAAAVCFEFQVE 82
PS L L K I LA+ AA CF F +E
Sbjct 477 LPSLLSVLGKHLLIANLASEAAECFRFVLE 506
> tpv:TP03_0199 DNA-directed RNA polymerase subunit alpha (EC:2.7.7.6);
K03040 DNA-directed RNA polymerase subunit alpha [EC:2.7.7.6]
Length=377
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query 1 NGALSVPGMREDVLELALNLKTLSLRS---GAPFARCRV-RVHARGPLKLLGGHISF 53
N SV G+RED EL NL+ + ++ A F + + +GP+ + GH+ F
Sbjct 3 NEFYSVVGLREDFFELVRNLRGVIFKNVPKTATFTNPVIATLRLKGPIIAVAGHLKF 59
> ath:AT5G15480 zinc finger (C2H2 type) family protein
Length=402
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 33 RCRVRVHARGPLKLLGGHISFPSFLKCLNKENYI 66
RCR+ + LGGH +F KC NK+N I
Sbjct 359 RCRLCNKIFSSYQALGGHQTFHRMSKCKNKKNGI 392
Lambda K H
0.326 0.140 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2035463248
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40