bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5792_orf4
Length=69
Score E
Sequences producing significant alignments: (Bits) Value
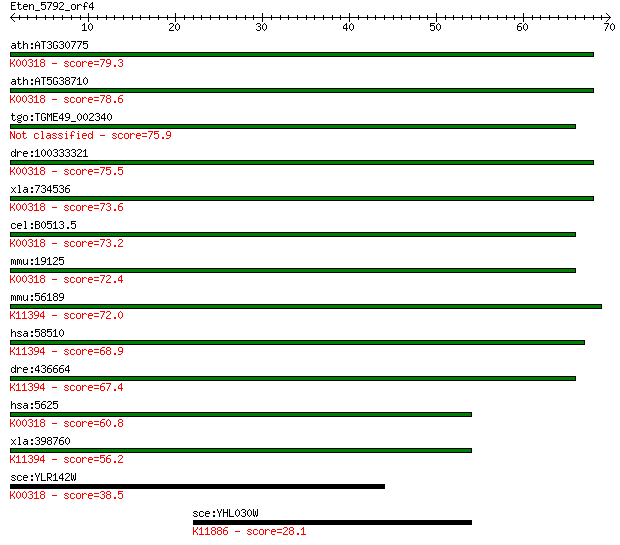
ath:AT3G30775 ERD5; ERD5 (EARLY RESPONSIVE TO DEHYDRATION 5); ... 79.3 3e-15
ath:AT5G38710 proline oxidase, putative / osmotic stress-respo... 78.6 4e-15
tgo:TGME49_002340 proline oxidase, putative (EC:1.5.99.8) 75.9 3e-14
dre:100333321 prodha, im:7136007, wu:fc02a12; proline dehydrog... 75.5 4e-14
xla:734536 prodh, MGC115247; proline dehydrogenase (oxidase) 1... 73.6 1e-13
cel:B0513.5 hypothetical protein; K00318 proline dehydrogenase... 73.2 2e-13
mmu:19125 Prodh, MGC159030, Pro-1, Pro1, Ym24d07; proline dehy... 72.4 3e-13
mmu:56189 Prodh2, 2510028N04Rik, 2510038B11Rik, MmPOX, MmPOX1,... 72.0 4e-13
hsa:58510 PRODH2, HSPOX1; proline dehydrogenase (oxidase) 2 (E... 68.9 4e-12
dre:436664 wu:fb24e01; zgc:92040 (EC:1.5.99.8); K11394 hydroxy... 67.4 1e-11
hsa:5625 PRODH, FLJ33744, HSPOX2, MGC148078, MGC148079, PIG6, ... 60.8 1e-09
xla:398760 prodh2, MGC68483; proline dehydrogenase (oxidase) 2... 56.2 3e-08
sce:YLR142W PUT1; Proline oxidase, nuclear-encoded mitochondri... 38.5 0.005
sce:YHL030W ECM29; Ecm29p; K11886 proteasome component ECM29 28.1 6.8
> ath:AT3G30775 ERD5; ERD5 (EARLY RESPONSIVE TO DEHYDRATION 5);
proline dehydrogenase (EC:1.5.99.8); K00318 proline dehydrogenase
[EC:1.5.99.8]
Length=499
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 36/67 (53%), Positives = 46/67 (68%), Gaps = 0/67 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QLYGM DA S GL AGF+V KY+PFGPV IPYLLRRA EN GM+ ++ + + E
Sbjct 430 QLYGMSDALSFGLKRAGFNVSKYMPFGPVATAIPYLLRRAYENRGMMATGAHDRQLMRME 489
Query 61 ITRRVLG 67
+ RR++
Sbjct 490 LKRRLIA 496
> ath:AT5G38710 proline oxidase, putative / osmotic stress-responsive
proline dehydrogenase, putative (EC:1.5.99.8); K00318
proline dehydrogenase [EC:1.5.99.8]
Length=476
Score = 78.6 bits (192), Expect = 4e-15, Method: Composition-based stats.
Identities = 35/67 (52%), Positives = 46/67 (68%), Gaps = 0/67 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QLYGM DA S GL AGF+V KY+P+GPV+ IPYL+RRA EN GM+ + + + E
Sbjct 409 QLYGMSDALSFGLKRAGFNVSKYMPYGPVDTAIPYLIRRAYENRGMMSTGALDRQLMRKE 468
Query 61 ITRRVLG 67
+ RRV+
Sbjct 469 LKRRVMA 475
> tgo:TGME49_002340 proline oxidase, putative (EC:1.5.99.8)
Length=485
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 36/65 (55%), Positives = 46/65 (70%), Gaps = 0/65 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL G+ D + LSS+GF VYKYVP+GPV TIPYLLRR +EN+G++G A E+ L E
Sbjct 413 QLLGLSDDLTFMLSSSGFKVYKYVPYGPVNVTIPYLLRRVQENSGIMGRAGAELVILYKE 472
Query 61 ITRRV 65
I R+
Sbjct 473 IKHRL 477
> dre:100333321 prodha, im:7136007, wu:fc02a12; proline dehydrogenase
(oxidase) 1a; K00318 proline dehydrogenase [EC:1.5.99.8]
Length=617
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 37/67 (55%), Positives = 43/67 (64%), Gaps = 0/67 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL GM D S L AGF VYKYVP+GPV + IPYL RRA+EN G + G+Q E L E
Sbjct 542 QLLGMCDQISFPLGQAGFPVYKYVPYGPVNEVIPYLSRRAQENRGFMKGSQRERSLLWKE 601
Query 61 ITRRVLG 67
I RR+
Sbjct 602 IKRRLFS 608
> xla:734536 prodh, MGC115247; proline dehydrogenase (oxidase)
1; K00318 proline dehydrogenase [EC:1.5.99.8]
Length=617
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/67 (53%), Positives = 44/67 (65%), Gaps = 0/67 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL GM D S L AG+ VYKYVP+GPV + +PYL RRA+EN G++ GA E R L E
Sbjct 542 QLLGMCDQISFPLGQAGYPVYKYVPYGPVHEVLPYLSRRAQENRGIMKGAIKERRLLWSE 601
Query 61 ITRRVLG 67
RR+L
Sbjct 602 FKRRLLS 608
> cel:B0513.5 hypothetical protein; K00318 proline dehydrogenase
[EC:1.5.99.8]
Length=616
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 35/65 (53%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QLYGM D S L AGF VYKY+P+GPVE+ +PYL RRA EN +L A E L E
Sbjct 539 QLYGMCDQVSFSLGQAGFSVYKYLPYGPVEEVLPYLSRRALENGSVLKKANKERDLLWKE 598
Query 61 ITRRV 65
+ RR+
Sbjct 599 LKRRI 603
> mmu:19125 Prodh, MGC159030, Pro-1, Pro1, Ym24d07; proline dehydrogenase
(EC:1.5.99.8); K00318 proline dehydrogenase [EC:1.5.99.8]
Length=599
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 35/65 (53%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL GM D S L AGF VYKYVP+GPV + +PYL RRA EN+ ++ GAQ E + L E
Sbjct 525 QLLGMCDQISFPLGQAGFPVYKYVPYGPVMEVLPYLSRRALENSSIMKGAQRERQLLWQE 584
Query 61 ITRRV 65
+ RR+
Sbjct 585 LRRRL 589
> mmu:56189 Prodh2, 2510028N04Rik, 2510038B11Rik, MmPOX, MmPOX1,
POX1; proline dehydrogenase (oxidase) 2 (EC:1.5.99.8); K11394
hydroxyproline oxidase [EC:1.5.-.-]
Length=456
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/68 (51%), Positives = 46/68 (67%), Gaps = 0/68 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL GM D S L AG+ VYK +P+G +E+ IPYL+RRA+EN +L GA+ E L E
Sbjct 387 QLLGMCDHVSLALGQAGYMVYKSIPYGCLEEVIPYLIRRAQENRSVLQGARREQALLSQE 446
Query 61 ITRRVLGR 68
+ RR+LGR
Sbjct 447 LWRRLLGR 454
> hsa:58510 PRODH2, HSPOX1; proline dehydrogenase (oxidase) 2
(EC:1.5.99.8); K11394 hydroxyproline oxidase [EC:1.5.-.-]
Length=536
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 33/66 (50%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL GM D S L AG+ VYK +P+G +E+ IPYL+RRA+EN +L GA+ E L E
Sbjct 463 QLLGMCDHVSLALGQAGYVVYKSIPYGSLEEVIPYLIRRAQENRSVLQGARREQELLSQE 522
Query 61 ITRRVL 66
+ RR+L
Sbjct 523 LWRRLL 528
> dre:436664 wu:fb24e01; zgc:92040 (EC:1.5.99.8); K11394 hydroxyproline
oxidase [EC:1.5.-.-]
Length=465
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 33/65 (50%), Positives = 43/65 (66%), Gaps = 0/65 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNEVRYLCHE 60
QL GM D S L+ GF VYK VP+G V+DT+PYL+RRA+EN +L G + E L E
Sbjct 376 QLLGMCDHVSLTLAQHGFSVYKSVPYGSVDDTLPYLVRRAQENRTVLQGIRKERDLLRQE 435
Query 61 ITRRV 65
+ RR+
Sbjct 436 LHRRL 440
> hsa:5625 PRODH, FLJ33744, HSPOX2, MGC148078, MGC148079, PIG6,
POX, PRODH1, PRODH2, SCZD4, TP53I6; proline dehydrogenase
(oxidase) 1 (EC:1.5.99.8); K00318 proline dehydrogenase [EC:1.5.99.8]
Length=492
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/53 (52%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNE 53
QL GM D S L AG+ VYKYVP+GPV + +PYL RRA EN+ ++ G E
Sbjct 418 QLLGMCDQISFPLGQAGYPVYKYVPYGPVMEVLPYLSRRALENSSLMKGTHRE 470
> xla:398760 prodh2, MGC68483; proline dehydrogenase (oxidase)
2 (EC:1.5.99.8); K11394 hydroxyproline oxidase [EC:1.5.-.-]
Length=466
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/53 (49%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 1 QLYGMGDAFSQGLSSAGFDVYKYVPFGPVEDTIPYLLRRARENTGMLGGAQNE 53
QL GM D S L AG+ VYK +P+G V+ +PYL+RRA+EN +L G + E
Sbjct 398 QLLGMCDHVSLTLGQAGYLVYKSLPYGSVDSVLPYLIRRAQENQSVLQGIRKE 450
> sce:YLR142W PUT1; Proline oxidase, nuclear-encoded mitochondrial
protein involved in utilization of proline as sole nitrogen
source; PUT1 transcription is induced by Put3p in the presence
of proline and the absence of a preferred nitrogen source
(EC:1.5.99.8); K00318 proline dehydrogenase [EC:1.5.99.8]
Length=476
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 20/45 (44%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 1 QLYGMGDAFSQGL--SSAGFDVYKYVPFGPVEDTIPYLLRRAREN 43
QL GM D + L + ++ KYVP+GP +T YLLRR +EN
Sbjct 406 QLLGMADNVTYDLITNHGAKNIIKYVPWGPPLETKDYLLRRLQEN 450
> sce:YHL030W ECM29; Ecm29p; K11886 proteasome component ECM29
Length=1868
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 10/32 (31%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 22 KYVPFGPVEDTIPYLLRRARENTGMLGGAQNE 53
+Y+ P E+TI +L+ +A+E +LG ++
Sbjct 1660 RYINVNPQEETITFLIEKAKEMIRLLGSESDD 1691
Lambda K H
0.323 0.143 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2029389012
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40