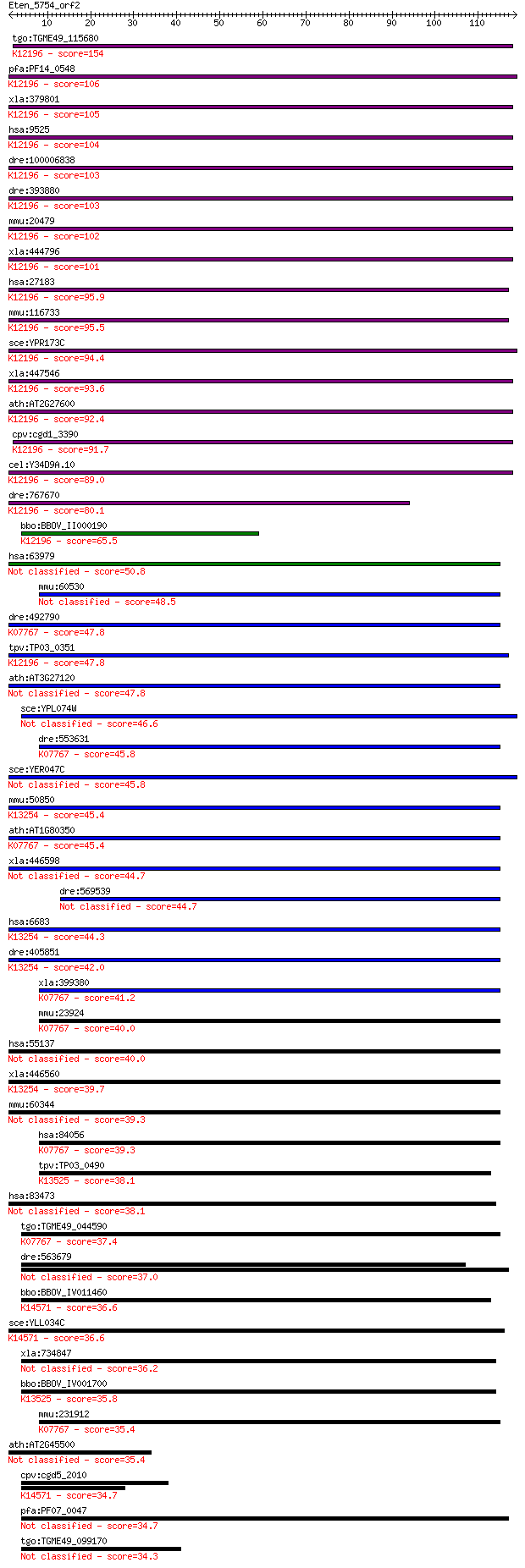
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5754_orf2
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12... 154 8e-38
pfa:PF14_0548 ATPase, putative; K12196 vacuolar protein-sortin... 106 2e-23
xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog... 105 3e-23
hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting ... 104 7e-23
dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog ... 103 1e-22
dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sortin... 103 1e-22
mmu:20479 Vps4b, 8030489C12Rik, Skd1; vacuolar protein sorting... 102 2e-22
xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-a... 101 5e-22
hsa:27183 VPS4A, FLJ22197, SKD1, SKD1A, SKD2, VPS4, VPS4-1; va... 95.9 3e-20
mmu:116733 Vps4a, 4930589C15Rik, AI325971, AW553189, MGC11698;... 95.5 4e-20
sce:YPR173C VPS4, CSC1, DID6, END13, GRD13, VPL4, VPT10; AAA-A... 94.4 8e-20
xla:447546 vps4a, vsp4; vacuolar protein sorting 4 homolog A; ... 93.6 2e-19
ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH DE... 92.4 3e-19
cpv:cgd1_3390 katanin p60/fidgetin family AAA ATpase ; K12196 ... 91.7 5e-19
cel:Y34D9A.10 vps-4; related to yeast Vacuolar Protein Sorting... 89.0 3e-18
dre:767670 vps4a, MGC153907, zgc:153907; vacuolar protein sort... 80.1 2e-15
bbo:BBOV_II000190 18.m05996; ATPase, AAA family; K12196 vacuol... 65.5 4e-11
hsa:63979 FIGNL1; fidgetin-like 1 50.8
mmu:60530 Fignl1; fidgetin-like 1 48.5
dre:492790 katnal1, zgc:101696; katanin p60 subunit A-like 1 (... 47.8 8e-06
tpv:TP03_0351 vacuolar sorting protein 4; K12196 vacuolar prot... 47.8 9e-06
ath:AT3G27120 ATP binding / ATPase/ nucleoside-triphosphatase/... 47.8 9e-06
sce:YPL074W YTA6; Putative ATPase of the CDC48/PAS1/SEC18 (AAA... 46.6 2e-05
dre:553631 katna1, MGC110580, si:dkey-15j16.1, zgc:110580; kat... 45.8 4e-05
sce:YER047C SAP1; Putative ATPase of the AAA family, interacts... 45.8 4e-05
mmu:50850 Spast, Spg4, mKIAA1083; spastin (EC:3.6.4.3); K13254... 45.4 4e-05
ath:AT1G80350 ERH3; ERH3 (ECTOPIC ROOT HAIR 3); ATP binding / ... 45.4 5e-05
xla:446598 fignl1, MGC81753; fidgetin-like 1 44.7
dre:569539 fignl1, fb82h05, wu:fb82h05, wu:fj99a11, zgc:193664... 44.7 8e-05
hsa:6683 SPAST, ADPSP, FSP2, KIAA1083, SPG4; spastin (EC:3.6.4... 44.3 1e-04
dre:405851 spast, MGC85952, zgc:85952; spastin (EC:3.6.4.3); K... 42.0 5e-04
xla:399380 katna1; katanin p60 (ATPase containing) subunit A 1... 41.2 0.001
mmu:23924 Katna1; katanin p60 (ATPase-containing) subunit A1 (... 40.0 0.002
hsa:55137 FIGN; fidgetin 40.0 0.002
xla:446560 spast, MGC81331, spg4; spastin (EC:3.6.4.3); K13254... 39.7 0.002
mmu:60344 Fign, fi, fidget; fidgetin 39.3 0.003
hsa:84056 KATNAL1, MGC2599; katanin p60 subunit A-like 1 (EC:3... 39.3 0.003
tpv:TP03_0490 cell division cycle protein 48; K13525 transitio... 38.1 0.007
hsa:83473 KATNAL2, DKFZp667C165, MGC33211; katanin p60 subunit... 38.1 0.008
tgo:TGME49_044590 p60 katanin, putative ; K07767 microtubule-s... 37.4 0.011
dre:563679 MGC136908; zgc:136908 37.0 0.016
bbo:BBOV_IV011460 23.m06332; ATPase AAA type domain containing... 36.6 0.020
sce:YLL034C RIX7; Putative ATPase of the AAA family, required ... 36.6 0.022
xla:734847 katnal2, MGC131314; katanin p60 subunit A-like 2 (E... 36.2 0.023
bbo:BBOV_IV001700 21.m02769; cell division cycle protein ATPas... 35.8 0.032
mmu:231912 Katnal1, MGC40859; katanin p60 subunit A-like 1 (EC... 35.4 0.039
ath:AT2G45500 ATP binding / nucleoside-triphosphatase/ nucleot... 35.4 0.044
cpv:cgd5_2010 nuclear VCP like protein with 2 AAA ATpase domai... 34.7 0.073
pfa:PF07_0047 cell division cycle ATPase, putative 34.7 0.078
tgo:TGME49_099170 ATPase, AAA family domain-containing protein 34.3 0.090
> tgo:TGME49_115680 vacuolar sorting ATPase Vps4, putative ; K12196
vacuolar protein-sorting-associated protein 4
Length=502
Score = 154 bits (388), Expect = 8e-38, Method: Compositional matrix adjust.
Identities = 70/116 (60%), Positives = 91/116 (78%), Gaps = 0/116 (0%)
Query 2 EGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSD 61
+ +F LAR+ EGFSG+DISV+VRDALF+P+R+CR AT FK+V ++G F PC PGDSD
Sbjct 386 DAEFDTLARQTEGFSGADISVVVRDALFQPLRKCRAATHFKRVFLDGTHFLSPCPPGDSD 445
Query 62 PTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGLEG 117
P+ +M LM VP +RLLPPE++MEDF + LR+ RPSVS ED+R H WT +FG+EG
Sbjct 446 PSKVEMRLMEVPPNRLLPPELSMEDFIAVLRNARPSVSEEDIRRHEEWTRRFGVEG 501
> pfa:PF14_0548 ATPase, putative; K12196 vacuolar protein-sorting-associated
protein 4
Length=419
Score = 106 bits (264), Expect = 2e-23, Method: Composition-based stats.
Identities = 44/118 (37%), Positives = 76/118 (64%), Gaps = 0/118 (0%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
S+ D + A E ++G+DI +L RDA++ P+++C + FKQV N + PC PGDS
Sbjct 302 SKEDIKQFATLTENYTGADIDILCRDAVYMPVKKCLLSKFFKQVKKNNKICYTPCSPGDS 361
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGLEGS 118
DPT + +MS+ + L P +T++DF +A+ + +PS+S +D++ + WT Q+G+ G+
Sbjct 362 DPTKVEKNVMSLSENELSLPPLTVQDFKTAISNAKPSLSVDDIKKYEEWTHQYGMNGT 419
> xla:379801 vps4b, MGC53483; vacuolar protein sorting 4 homolog
B; K12196 vacuolar protein-sorting-associated protein 4
Length=442
Score = 105 bits (263), Expect = 3e-23, Method: Composition-based stats.
Identities = 55/125 (44%), Positives = 75/125 (60%), Gaps = 10/125 (8%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVM--------INGCAFF 52
SE DFR L ++ G+SG+DIS++VRDAL +P+R+ ++AT FK+V +
Sbjct 320 SEPDFRDLGKKTNGYSGADISIIVRDALMQPVRKVQSATHFKRVKGKSPLDPNVTRDDLL 379
Query 53 VPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
PC PG DP A +M M VP D+L P V M D +L HT+P+V+ EDL +TE
Sbjct 380 TPCSPG--DPNAVEMTWMDVPGDKLFEPVVCMSDMLKSLAHTKPTVNDEDLTKLKKFTED 437
Query 113 FGLEG 117
FG EG
Sbjct 438 FGQEG 442
> hsa:9525 VPS4B, SKD1, SKD1B, VPS4-2; vacuolar protein sorting
4 homolog B (S. cerevisiae); K12196 vacuolar protein-sorting-associated
protein 4
Length=444
Score = 104 bits (260), Expect = 7e-23, Method: Composition-based stats.
Identities = 55/125 (44%), Positives = 77/125 (61%), Gaps = 10/125 (8%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCA--------FF 52
+E DFR L R+ +G+SG+DIS++VRDAL +P+R+ ++AT FK+V A
Sbjct 322 TEADFRELGRKTDGYSGADISIIVRDALMQPVRKVQSATHFKKVRGPSRADPNHLVDDLL 381
Query 53 VPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
PC PG DP A +M M VP D+LL P V+M D +L +T+P+V+ DL +TE
Sbjct 382 TPCSPG--DPGAIEMTWMDVPGDKLLEPVVSMSDMLRSLSNTKPTVNEHDLLKLKKFTED 439
Query 113 FGLEG 117
FG EG
Sbjct 440 FGQEG 444
> dre:100006838 ch73-81e6.1; vacuolar protein sorting 4 homolog
b-like; K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 52/125 (41%), Positives = 78/125 (62%), Gaps = 10/125 (8%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCA--------FF 52
+E DF L ++ +G+SG+DIS++VRDAL +P+R+ ++AT FKQV +
Sbjct 315 TESDFMTLGKKTDGYSGADISIIVRDALMQPVRKVQSATHFKQVRGPSRSDPNVIVDDLL 374
Query 53 VPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
PC PG DP A++M M VP ++LL P V+M D +L +T+P+V+ +DL +TE
Sbjct 375 TPCSPG--DPQAKEMTWMEVPGEKLLEPIVSMSDMLRSLSNTKPTVNEQDLEKLKKFTED 432
Query 113 FGLEG 117
FG EG
Sbjct 433 FGQEG 437
> dre:393880 vps4b, MGC63682, zgc:63682; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=437
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 52/125 (41%), Positives = 78/125 (62%), Gaps = 10/125 (8%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCA--------FF 52
+E DF L ++ +G+SG+DIS++VRDAL +P+R+ ++AT FKQV +
Sbjct 315 TESDFMTLGKKTDGYSGADISIIVRDALMQPVRKVQSATHFKQVRGPSRSDPNVIVDDLL 374
Query 53 VPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
PC PG DP A++M M VP ++LL P V+M D +L +T+P+V+ +DL +TE
Sbjct 375 TPCSPG--DPQAKEMTWMEVPGEKLLEPIVSMSDMLRSLSNTKPTVNEQDLEKLKKFTED 432
Query 113 FGLEG 117
FG EG
Sbjct 433 FGQEG 437
> mmu:20479 Vps4b, 8030489C12Rik, Skd1; vacuolar protein sorting
4b (yeast); K12196 vacuolar protein-sorting-associated protein
4
Length=444
Score = 102 bits (255), Expect = 2e-22, Method: Composition-based stats.
Identities = 54/127 (42%), Positives = 79/127 (62%), Gaps = 14/127 (11%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQV----------MINGCA 50
+E DF+ L R+ +G+SG+DIS++VRDAL +P+R+ ++AT FK+V ++N
Sbjct 322 TEADFQELGRKTDGYSGADISIIVRDALMQPVRKVQSATHFKKVRGPSRADPNCIVND-- 379
Query 51 FFVPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWT 110
PC PG DP A +M M VP D+LL P V+M D +L T+P+V+ +DL +T
Sbjct 380 LLTPCSPG--DPGAIEMTWMDVPGDKLLEPVVSMWDMLRSLSSTKPTVNEQDLLKLKKFT 437
Query 111 EQFGLEG 117
E FG EG
Sbjct 438 EDFGQEG 444
> xla:444796 MGC82073 protein; K12196 vacuolar protein-sorting-associated
protein 4
Length=443
Score = 101 bits (252), Expect = 5e-22, Method: Composition-based stats.
Identities = 54/125 (43%), Positives = 75/125 (60%), Gaps = 10/125 (8%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVM--------INGCAFF 52
SE DFR L ++ G+SG+DIS++VRDAL +P+R+ ++AT FK+ +
Sbjct 321 SEPDFRDLGKKTNGYSGADISIIVRDALMQPVRKVQSATHFKKERGKSPLDPNVTRDDLL 380
Query 53 VPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
PC PGD P A +M + VP D+LL P V M D +L HT+P+V+ EDL +TE
Sbjct 381 TPCSPGD--PNAVEMTWVDVPGDKLLEPVVCMPDMLKSLAHTKPTVNDEDLAKLRKFTED 438
Query 113 FGLEG 117
FG EG
Sbjct 439 FGQEG 443
> hsa:27183 VPS4A, FLJ22197, SKD1, SKD1A, SKD2, VPS4, VPS4-1;
vacuolar protein sorting 4 homolog A (S. cerevisiae); K12196
vacuolar protein-sorting-associated protein 4
Length=437
Score = 95.9 bits (237), Expect = 3e-20, Method: Composition-based stats.
Identities = 52/126 (41%), Positives = 76/126 (60%), Gaps = 14/126 (11%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQV----------MINGCA 50
++ + LAR+ EG+SG+DIS++VRD+L +P+R+ ++AT FK+V MI+
Sbjct 315 TDANIHELARKTEGYSGADISIIVRDSLMQPVRKVQSATHFKKVCGPSRTNPSMMIDD-- 372
Query 51 FFVPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWT 110
PC PG DP A +M M VP D+LL P V M D +L TRP+V+ +DL ++
Sbjct 373 LLTPCSPG--DPGAMEMTWMDVPGDKLLEPVVCMSDMLRSLATTRPTVNADDLLKVKKFS 430
Query 111 EQFGLE 116
E FG E
Sbjct 431 EDFGQE 436
> mmu:116733 Vps4a, 4930589C15Rik, AI325971, AW553189, MGC11698;
vacuolar protein sorting 4a (yeast); K12196 vacuolar protein-sorting-associated
protein 4
Length=437
Score = 95.5 bits (236), Expect = 4e-20, Method: Composition-based stats.
Identities = 52/126 (41%), Positives = 76/126 (60%), Gaps = 14/126 (11%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQ----------VMINGCA 50
++ + LAR+ EG+SG+DIS++VRD+L +P+R+ ++AT FK+ VMI+
Sbjct 315 TDANIHELARKTEGYSGADISIIVRDSLMQPVRKVQSATHFKKVCGPSRTNPSVMIDD-- 372
Query 51 FFVPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWT 110
PC PG DP A +M M VP D+LL P V M D +L TRP+V+ +DL ++
Sbjct 373 LLTPCSPG--DPGAIEMTWMDVPGDKLLEPVVCMSDMLRSLATTRPTVNADDLLKVKKFS 430
Query 111 EQFGLE 116
E FG E
Sbjct 431 EDFGQE 436
> sce:YPR173C VPS4, CSC1, DID6, END13, GRD13, VPL4, VPT10; AAA-ATPase
involved in multivesicular body (MVB) protein sorting,
ATP-bound Vps4p localizes to endosomes and catalyzes ESCRT-III
disassembly and membrane release; ATPase activity is activated
by Vta1p; regulates cellular sterol metabolism; K12196
vacuolar protein-sorting-associated protein 4
Length=437
Score = 94.4 bits (233), Expect = 8e-20, Method: Composition-based stats.
Identities = 49/120 (40%), Positives = 75/120 (62%), Gaps = 4/120 (3%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMI--NGCAFFVPCHPG 58
++ D+R L EG+SGSDI+V+V+DAL +PIR+ ++AT FK V + PC PG
Sbjct 320 TKEDYRTLGAMTEGYSGSDIAVVVKDALMQPIRKIQSATHFKDVSTEDDETRKLTPCSPG 379
Query 59 DSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGLEGS 118
D A +M+ + AD L P++T++DF A++ TRP+V+ +DL +T FG EG+
Sbjct 380 DDG--AIEMSWTDIEADELKEPDLTIKDFLKAIKSTRPTVNEDDLLKQEQFTRDFGQEGN 437
> xla:447546 vps4a, vsp4; vacuolar protein sorting 4 homolog A;
K12196 vacuolar protein-sorting-associated protein 4
Length=436
Score = 93.6 bits (231), Expect = 2e-19, Method: Composition-based stats.
Identities = 52/125 (41%), Positives = 75/125 (60%), Gaps = 10/125 (8%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVM--------INGCAFF 52
SE + R LA++ +G+SG+DIS++VRDAL +P+R+ ++AT FK+V I
Sbjct 314 SEENVRELAKKTDGYSGADISIIVRDALMQPVRKVQSATHFKKVRGPSRTNPGIIVDDLL 373
Query 53 VPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
PC PG DP A +M M V +D+L P V M D +L TRP+V+ +DL +T+
Sbjct 374 TPCSPG--DPGAVEMTWMEVSSDKLQEPVVCMSDMLRSLATTRPTVNADDLLKVKKFTDD 431
Query 113 FGLEG 117
FG EG
Sbjct 432 FGQEG 436
> ath:AT2G27600 SKD1; SKD1 (SUPPRESSOR OF K+ TRANSPORT GROWTH
DEFECT1); ATP binding / nucleoside-triphosphatase/ nucleotide
binding; K12196 vacuolar protein-sorting-associated protein
4
Length=435
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 49/120 (40%), Positives = 73/120 (60%), Gaps = 7/120 (5%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPC---HP 57
+E DF YL ++ EGFSGSD+SV V+D LFEP+R+ + A F + ++PC HP
Sbjct 320 TEPDFEYLGQKTEGFSGSDVSVCVKDVLFEPVRKTQDAMFF---FKSPDGTWMPCGPRHP 376
Query 58 GDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGLEG 117
G T + +A + A++++PP +T DF L RP+VS DL +H +T++FG EG
Sbjct 377 GAIQTTMQDLATKGL-AEKIIPPPITRTDFEKVLARQRPTVSKSDLDVHERFTQEFGEEG 435
> cpv:cgd1_3390 katanin p60/fidgetin family AAA ATpase ; K12196
vacuolar protein-sorting-associated protein 4
Length=462
Score = 91.7 bits (226), Expect = 5e-19, Method: Composition-based stats.
Identities = 47/134 (35%), Positives = 73/134 (54%), Gaps = 18/134 (13%)
Query 2 EGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMI-------------NG 48
+ D Y+A+ G+S SD+S+L++DALFEPIR+C + FK+V+I N
Sbjct 328 DDDINYIAKMTHGYSSSDVSILIKDALFEPIRKCSESNWFKKVVIMNNNDEITNNNAENF 387
Query 49 CAFFVPC-HPGDSD----PTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDL 103
++ PC P + D RK +L +P ++LLPP++T D L T+ S++ D+
Sbjct 388 KIYWTPCSQPSNIDHYDKELYRKTSLYDIPNNQLLPPKLTKSDLIHVLSKTKSSITNLDI 447
Query 104 RMHISWTEQFGLEG 117
WT +FGL G
Sbjct 448 DKFTEWTNKFGLSG 461
> cel:Y34D9A.10 vps-4; related to yeast Vacuolar Protein Sorting
factor family member (vps-4); K12196 vacuolar protein-sorting-associated
protein 4
Length=430
Score = 89.0 bits (219), Expect = 3e-18, Method: Composition-based stats.
Identities = 47/125 (37%), Positives = 70/125 (56%), Gaps = 10/125 (8%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVM--------INGCAFF 52
++ DF+ LA EG+SG DIS+LV+DAL +P+RR ++AT FK V +
Sbjct 306 TDQDFKVLAERCEGYSGYDISILVKDALMQPVRRVQSATHFKHVSGPSPKDPNVIAHDLL 365
Query 53 VPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
PC PG DP A M + VP D+L P ++M+D +L +P+V+ DL ++
Sbjct 366 TPCSPG--DPHAIAMNWLDVPGDKLANPPLSMQDISRSLASVKPTVNNTDLDRLEAFKND 423
Query 113 FGLEG 117
FG +G
Sbjct 424 FGQDG 428
> dre:767670 vps4a, MGC153907, zgc:153907; vacuolar protein sorting
4a (yeast); K12196 vacuolar protein-sorting-associated
protein 4
Length=440
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 46/104 (44%), Positives = 62/104 (59%), Gaps = 13/104 (12%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQV----MINGCA----FF 52
+E D R LAR+ +G+SG+DIS++VRDAL +P+R+ ++AT FK+V N
Sbjct 314 TEADLRQLARKTDGYSGADISIIVRDALMQPVRKVQSATHFKKVRGPSRSNSAVIVDDLL 373
Query 53 VPCHPGDSDPTARKMALMSVPADRLLPPEVTMEDF---HSALRH 93
PC PG DP A +M M VP D+LL P V M + SAL H
Sbjct 374 TPCSPG--DPEAIEMTWMDVPGDKLLEPIVCMVRYRGSRSALLH 415
> bbo:BBOV_II000190 18.m05996; ATPase, AAA family; K12196 vacuolar
protein-sorting-associated protein 4
Length=363
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 28/55 (50%), Positives = 40/55 (72%), Gaps = 0/55 (0%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPG 58
D LA+ EG+SGSD++V+VRDA +P+R+CR A+ FK+V+ NG F+ PC G
Sbjct 306 DLDELAQCTEGYSGSDVNVVVRDARMQPLRKCRDASFFKKVIRNGEEFYTPCAAG 360
> hsa:63979 FIGNL1; fidgetin-like 1
Length=674
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 31/114 (27%), Positives = 53/114 (46%), Gaps = 31/114 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
SE + + ++ + FSG+D++ L R+A PIR +TA
Sbjct 589 SEEEIEQIVQQSDAFSGADMTQLCREASLGPIRSLQTAD--------------------- 627
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ ++ D++ P + DF +A R RPSVSP+DL ++ +W + FG
Sbjct 628 --------IATITPDQVRP--IAYIDFENAFRTVRPSVSPKDLELYENWNKTFG 671
> mmu:60530 Fignl1; fidgetin-like 1
Length=683
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 50/107 (46%), Gaps = 31/107 (28%)
Query 8 LARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPTARKM 67
+ ++ +GFSG+D++ L R+A PIR H D
Sbjct 605 VVQQSDGFSGADMTQLCREASLGPIRSL--------------------HAAD-------- 636
Query 68 ALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ ++ D++ P + DF +A + RP+VSP+DL ++ +W E FG
Sbjct 637 -IATISPDQVRP--IAYIDFENAFKTVRPTVSPKDLELYENWNETFG 680
> dre:492790 katnal1, zgc:101696; katanin p60 subunit A-like 1
(EC:3.6.4.3); K07767 microtubule-severing ATPase [EC:3.6.4.3]
Length=488
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 53/114 (46%), Gaps = 26/114 (22%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
S+ D A ++EG+SG+DI+ + RDA +RR I G + P +
Sbjct 399 SDVDLTVFAEKIEGYSGADITNVCRDASMMAMRR----------RIQGLS------PEE- 441
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ ++ D L P VTMEDF AL+ SVS DL + SW +FG
Sbjct 442 --------IRALSKDELQMP-VTMEDFELALKKISKSVSAADLEKYESWMSEFG 486
> tpv:TP03_0351 vacuolar sorting protein 4; K12196 vacuolar protein-sorting-associated
protein 4
Length=362
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 23/116 (19%), Positives = 54/116 (46%), Gaps = 28/116 (24%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
++ D Y++ F+ DI++LV++ + +++
Sbjct 274 ADSDLHYMSENTTNFNCYDINILVKEIVLYTLKKYNN----------------------- 310
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGLE 116
+L+++ D L+ P +++ED L+ RPSV+ +D++++ WT+Q+G +
Sbjct 311 -----NESLLNINEDDLIIPTISIEDVKEVLKDFRPSVAVDDIKLYEQWTQQYGTQ 361
> ath:AT3G27120 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=476
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 51/114 (44%), Gaps = 29/114 (25%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
S+ D + EG+SGSD+ LV+DA P+R
Sbjct 388 SDDDMNIICNLTEGYSGSDMKNLVKDATMGPLR--------------------------- 420
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ R + + ++ D + VT++DF AL+ RPSVS +L ++ +W QFG
Sbjct 421 EALKRGIDITNLTKDDM--RLVTLQDFKDALQEVRPSVSQNELGIYENWNNQFG 472
> sce:YPL074W YTA6; Putative ATPase of the CDC48/PAS1/SEC18 (AAA)
family, localized to the cortex of mother cells but not
to daughter cells
Length=754
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 34/115 (29%), Positives = 50/115 (43%), Gaps = 32/115 (27%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPT 63
D+ + EGFSGSD++ L ++A EPIR GD
Sbjct 672 DYELITEMTEGFSGSDLTSLAKEAAMEPIRDL----------------------GDK--- 706
Query 64 ARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGLEGS 118
LM D++ E+ +DF +AL + SVS E L+ + W+ +FG GS
Sbjct 707 -----LMFADFDKIRGIEI--KDFQNALLTIKKSVSSESLQKYEEWSSKFGSNGS 754
> dre:553631 katna1, MGC110580, si:dkey-15j16.1, zgc:110580; katanin
p60 (ATPase-containing) subunit A 1 (EC:3.6.4.3); K07767
microtubule-severing ATPase [EC:3.6.4.3]
Length=485
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 33/107 (30%), Positives = 50/107 (46%), Gaps = 26/107 (24%)
Query 8 LARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPTARKM 67
+A ++EG+SG+DI+ + RDA +RR I G P +
Sbjct 403 IAEQMEGYSGADITNVCRDASLMAMRR----------RIEGLT------PEE-------- 438
Query 68 ALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ ++P D + P TMEDF +AL+ SVS DL + W +FG
Sbjct 439 -IRNLPKDEMHMP-TTMEDFETALKKVSKSVSAADLEKYEKWIAEFG 483
> sce:YER047C SAP1; Putative ATPase of the AAA family, interacts
with the Sin1p transcriptional repressor in the two-hybrid
system
Length=897
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 34/118 (28%), Positives = 51/118 (43%), Gaps = 32/118 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
+E DF L + EG+SGSDI+ L +DA P+R GD
Sbjct 812 TESDFDELVKITEGYSGSDITSLAKDAAMGPLRDL----------------------GDK 849
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGLEGS 118
L+ + + P + + DF ++L + +PSVS + L + W QFG GS
Sbjct 850 --------LLETEREMIRP--IGLVDFKNSLVYIKPSVSQDGLVKYEKWASQFGSSGS 897
> mmu:50850 Spast, Spg4, mKIAA1083; spastin (EC:3.6.4.3); K13254
spastin [EC:3.6.4.3]
Length=614
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 48/114 (42%), Gaps = 31/114 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
++ + LAR +G+SGSD++ L +DA PIR +
Sbjct 528 TQKELAQLARMTDGYSGSDLTALAKDAALGPIRELK------------------------ 563
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
++ MS R + + DF +L+ + SVSP+ L +I W + FG
Sbjct 564 ---PEQVKNMSASEMR----NIRLSDFTESLKKIKRSVSPQTLEAYIRWNKDFG 610
> ath:AT1G80350 ERH3; ERH3 (ECTOPIC ROOT HAIR 3); ATP binding
/ nucleoside-triphosphatase/ nucleotide binding; K07767 microtubule-severing
ATPase [EC:3.6.4.3]
Length=523
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 47/114 (41%), Gaps = 25/114 (21%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
S+ + +AR EG+SG D++ + RDA +RR + A + + N + P
Sbjct 433 SDVNIEDVARRTEGYSGDDLTNVCRDASMNGMRR-KIAGKTRDEIKNMSKDDISNDP--- 488
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
V M DF A+R +PSVS D+ H W +FG
Sbjct 489 ---------------------VAMCDFEEAIRKVQPSVSSSDIEKHEKWLSEFG 521
> xla:446598 fignl1, MGC81753; fidgetin-like 1
Length=655
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 29/114 (25%), Positives = 52/114 (45%), Gaps = 31/114 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
+E + + + +GFSG+D++ L R+A PIR
Sbjct 570 TEQEVEAIVLQADGFSGADMTQLCREAALGPIR--------------------------- 602
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ + M + ++ A+++ P + DF SA RPSVS +DL ++ +W + FG
Sbjct 603 --SIQLMDISTITAEQVRP--IAYIDFQSAFLVVRPSVSQKDLELYENWNKTFG 652
> dre:569539 fignl1, fb82h05, wu:fb82h05, wu:fj99a11, zgc:193664;
fidgetin-like 1
Length=661
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 28/102 (27%), Positives = 45/102 (44%), Gaps = 31/102 (30%)
Query 13 EGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPTARKMALMSV 72
EGFSG+D++ L R+A PIR + + ++
Sbjct 588 EGFSGADMTQLCREAALGPIRSISLSD-----------------------------IATI 618
Query 73 PADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
A+++ P + DF AL+ RPSVS +DL ++ W + FG
Sbjct 619 MAEQVRP--ILYSDFQEALKTVRPSVSSKDLELYEEWNKTFG 658
> hsa:6683 SPAST, ADPSP, FSP2, KIAA1083, SPG4; spastin (EC:3.6.4.3);
K13254 spastin [EC:3.6.4.3]
Length=616
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 29/114 (25%), Positives = 48/114 (42%), Gaps = 31/114 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
++ + LAR +G+SGSD++ L +DA PIR +
Sbjct 530 TQKELAQLARMTDGYSGSDLTALAKDAALGPIRELK------------------------ 565
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
++ MS R + + DF +L+ + SVSP+ L +I W + FG
Sbjct 566 ---PEQVKNMSASEMR----NIRLSDFTESLKKIKRSVSPQTLEAYIRWNKDFG 612
> dre:405851 spast, MGC85952, zgc:85952; spastin (EC:3.6.4.3);
K13254 spastin [EC:3.6.4.3]
Length=570
Score = 42.0 bits (97), Expect = 5e-04, Method: Composition-based stats.
Identities = 29/114 (25%), Positives = 52/114 (45%), Gaps = 31/114 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
S+ + LAR +G+SGSD++ L +DA PIR + +QV
Sbjct 483 SQKELSQLARLTDGYSGSDLTSLAKDAALGPIRELKP----EQV---------------R 523
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ +A +M ++ + DF +L+ + SVSP+ L ++ W ++G
Sbjct 524 NMSAHEMR------------DIRISDFLESLKRIKRSVSPQTLDQYVRWNREYG 565
> xla:399380 katna1; katanin p60 (ATPase containing) subunit A
1 (EC:3.6.4.3); K07767 microtubule-severing ATPase [EC:3.6.4.3]
Length=488
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 26/107 (24%)
Query 8 LARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPTARKM 67
+A ++G+SG+DI+ + RDA +RR I G P + +R
Sbjct 406 IAENMDGYSGADITNVCRDASLMAMRR----------RIEGLT------PEEIRNLSRD- 448
Query 68 ALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
D +P TMEDF AL+ SVS D+ + W E+FG
Sbjct 449 -------DMHMP--TTMEDFEMALKKVSKSVSASDIEKYEKWIEEFG 486
> mmu:23924 Katna1; katanin p60 (ATPase-containing) subunit A1
(EC:3.6.4.3); K07767 microtubule-severing ATPase [EC:3.6.4.3]
Length=493
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 26/107 (24%)
Query 8 LARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPTARKM 67
+A +EG+SG+DI+ + RDA +RR I G P + +R+
Sbjct 411 IAENMEGYSGADITNVCRDASLMAMRR----------RIEGLT------PEEIRNLSREA 454
Query 68 ALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
M +P TMEDF AL+ SVS D+ + W +FG
Sbjct 455 --MHMP--------TTMEDFEMALKKISKSVSAADIERYEKWIVEFG 491
> hsa:55137 FIGN; fidgetin
Length=759
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 29/114 (25%), Positives = 49/114 (42%), Gaps = 31/114 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
++ +F L + EGFSG D++ L ++A+ P+ AT +M P
Sbjct 674 NDKEFALLVQRTEGFSGLDVAHLCQEAVVGPLH-AMPATDLSAIM-----------PSQL 721
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
P VT +DF +A +PS+S ++L M++ W + FG
Sbjct 722 RP-------------------VTYQDFENAFCKIQPSISQKELDMYVEWNKMFG 756
> xla:446560 spast, MGC81331, spg4; spastin (EC:3.6.4.3); K13254
spastin [EC:3.6.4.3]
Length=600
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 33/114 (28%), Positives = 47/114 (41%), Gaps = 31/114 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
SE + L+R EG+SGSDI+ L +DA PIR + +QV
Sbjct 514 SEKELTQLSRLTEGYSGSDITALAKDAALGPIRELKP----EQV---------------K 554
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ A +M M DF +L+ + SVS L +I W + FG
Sbjct 555 NMAASEMRNMK------------YSDFLGSLKKIKCSVSHSTLESYIRWNQDFG 596
> mmu:60344 Fign, fi, fidget; fidgetin
Length=759
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 29/114 (25%), Positives = 48/114 (42%), Gaps = 31/114 (27%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
++ +F L + EGFSG D++ L ++A P+ AT +M P
Sbjct 674 NDKEFALLVQRTEGFSGLDVAHLCQEAAVGPLH-AMPATDLSAIM-----------PSQL 721
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
P VT +DF +A +PS+S ++L M++ W + FG
Sbjct 722 RP-------------------VTYQDFENAFCKIQPSISQKELDMYVEWNKMFG 756
> hsa:84056 KATNAL1, MGC2599; katanin p60 subunit A-like 1 (EC:3.6.4.3);
K07767 microtubule-severing ATPase [EC:3.6.4.3]
Length=490
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 49/107 (45%), Gaps = 26/107 (24%)
Query 8 LARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPTARKM 67
+A ++EG+SG+DI+ + RDA +RR ING + P +
Sbjct 408 IAEKIEGYSGADITNVCRDASLMAMRR----------RINGLS------PEE-------- 443
Query 68 ALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ ++ + L P VT DF AL+ SVS DL + W +FG
Sbjct 444 -IRALSKEELQMP-VTKGDFELALKKIAKSVSAADLEKYEKWMVEFG 488
> tpv:TP03_0490 cell division cycle protein 48; K13525 transitional
endoplasmic reticulum ATPase
Length=954
Score = 38.1 bits (87), Expect = 0.007, Method: Composition-based stats.
Identities = 27/113 (23%), Positives = 51/113 (45%), Gaps = 41/113 (36%)
Query 8 LARELEGFSGSDISVL--------VRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGD 59
+A++L+G+SG+DI+ + +R+++ E I+R R P G+
Sbjct 872 MAQQLDGYSGADIAEICHRAAREAIRESIEEEIKRKR-----------------PLEKGE 914
Query 60 SDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
DP P +T + F ALR++R SV D++++ S+ +
Sbjct 915 KDPV----------------PFITNKHFQVALRNSRKSVEQSDIQLYESFKNK 951
> hsa:83473 KATNAL2, DKFZp667C165, MGC33211; katanin p60 subunit
A-like 2 (EC:3.6.4.3)
Length=466
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 31/113 (27%), Positives = 49/113 (43%), Gaps = 26/113 (23%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDS 60
+E ++ L++E EG+SGSDI ++ R+A P+R+ A + H +S
Sbjct 377 TELEYSVLSQETEGYSGSDIKLVCREAAMRPVRKIFDA--------------LENHQSES 422
Query 61 DPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQF 113
+P R+ VT DF L HT+PS R + W +F
Sbjct 423 S---------DLP--RIQLDIVTTADFLDVLTHTKPSAKNLAQR-YSDWQREF 463
> tgo:TGME49_044590 p60 katanin, putative ; K07767 microtubule-severing
ATPase [EC:3.6.4.3]
Length=410
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 41/111 (36%), Gaps = 22/111 (19%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPT 63
DF +A E FSG+D+ L R+A P+RR F + P D
Sbjct 320 DFLQIANRTEQFSGADLQHLCREACMNPLRRV-----FADL------------PLDEIKA 362
Query 64 ARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
R+ R V+M DF AL P+ ++ W +FG
Sbjct 363 KREAGAFGEEQTR-----VSMADFEQALEKANPATHAAEIAKFEKWNAEFG 408
> dre:563679 MGC136908; zgc:136908
Length=805
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 47/111 (42%), Gaps = 34/111 (30%)
Query 4 DFRYLARELEGFSGSDISVL--------VRDALFEPIRRCRTATAFKQVMINGCAFFVPC 55
D YL++ EGFSG+D++ + +R+A+ IR R A K+ ++
Sbjct 673 DLMYLSKITEGFSGADLTEICQRACKLAIREAIEAEIRAERQRQARKETAMDD------- 725
Query 56 HPGDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMH 106
D DP PE+ + F A+R R SVS D+R +
Sbjct 726 ---DYDPV----------------PEIRKDHFEEAMRFARRSVSDNDIRKY 757
Score = 33.1 bits (74), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 29/115 (25%), Positives = 48/115 (41%), Gaps = 28/115 (24%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPT 63
D ++ E G G+D++ L +A + IR+ K +I+
Sbjct 397 DLEQISAETHGHVGADLAALCSEAALQAIRK-------KMTLID---------------- 433
Query 64 ARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRM--HISWTEQFGLE 116
+ S+ AD L VTM+DF AL + PS E + H++W + GL+
Sbjct 434 ---LEDDSIDADLLNSLAVTMDDFKWALSQSNPSALRETVVEVPHVNWEDIGGLD 485
> bbo:BBOV_IV011460 23.m06332; ATPase AAA type domain containing
protein; K14571 ribosome biogenesis ATPase
Length=707
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 52/111 (46%), Gaps = 26/111 (23%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPT 63
DF+ +A + EG+SG+D++ LVR+A + + R ++ H D+
Sbjct 616 DFQKIAVQTEGYSGADLACLVREAGISAVEKLRIQ-------------YIKEHGLDT--- 659
Query 64 ARKMALMSVPADRLLPPE--VTMEDFHSALRHTRPSVSPEDLRMHISWTEQ 112
+ SV A PP ++ ED SAL PSV+ + + + S+ ++
Sbjct 660 ----YVRSVDA----PPGACISAEDLASALLKVSPSVTQKQINFYESFQQR 702
> sce:YLL034C RIX7; Putative ATPase of the AAA family, required
for export of pre-ribosomal large subunits from the nucleus;
distributed between the nucleolus, nucleoplasm, and nuclear
periphery depending on growth conditions; K14571 ribosome
biogenesis ATPase
Length=837
Score = 36.6 bits (83), Expect = 0.022, Method: Composition-based stats.
Identities = 32/117 (27%), Positives = 57/117 (48%), Gaps = 16/117 (13%)
Query 1 SEGDFRYLAR--ELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPG 58
S+ DF + R + FSG+D++ LVR++ ++R + Q +++
Sbjct 723 SDVDFEEIIRNEKCNNFSGADLAALVRESSVLALKRKFFQSEEIQSVLDN---------- 772
Query 59 DSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGL 115
D D +++ V + ++ VTM DF SALR +PSVS +D + ++ GL
Sbjct 773 DLDKEFEDLSV-GVSGEEII---VTMSDFRSALRKIKPSVSDKDRLKYDRLNKKMGL 825
> xla:734847 katnal2, MGC131314; katanin p60 subunit A-like 2
(EC:3.6.4.3)
Length=505
Score = 36.2 bits (82), Expect = 0.023, Method: Composition-based stats.
Identities = 26/110 (23%), Positives = 46/110 (41%), Gaps = 26/110 (23%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPT 63
D+ L E +G+SGSDI ++ ++A P+R+ A +
Sbjct 419 DYSTLGEETDGYSGSDIRLVCKEAAMRPVRKIFDALE------------------NHHSE 460
Query 64 ARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQF 113
+K+ ++S+ VT DF L HT+PS + + +W +F
Sbjct 461 HKKLPVISLET-------VTTSDFSEVLAHTKPSAKSLAEK-YSAWQNEF 502
> bbo:BBOV_IV001700 21.m02769; cell division cycle protein ATPase;
K13525 transitional endoplasmic reticulum ATPase
Length=922
Score = 35.8 bits (81), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 47/110 (42%), Gaps = 29/110 (26%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPT 63
+ R +A ELEG+SG+DI+ + A E IR + + + G G+ DP
Sbjct 833 NIRRMAEELEGYSGADIAEICHRAAREAIRE-----SIEHEIKRGRRL----KEGEEDPV 883
Query 64 ARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQF 113
P +T E F A+ + R SV ED++ + EQF
Sbjct 884 ----------------PYITNEHFRVAMANARKSVRKEDIKRY----EQF 913
> mmu:231912 Katnal1, MGC40859; katanin p60 subunit A-like 1 (EC:3.6.4.3);
K07767 microtubule-severing ATPase [EC:3.6.4.3]
Length=488
Score = 35.4 bits (80), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 47/107 (43%), Gaps = 26/107 (24%)
Query 8 LARELEGFSGSDISVLVRDALFEPIRRCRTATAFKQVMINGCAFFVPCHPGDSDPTARKM 67
+A + EG+SG+DI+ + RDA +RR ING + P +
Sbjct 406 IADKTEGYSGADITNICRDASLMAMRR----------RINGLS------PEE-------- 441
Query 68 ALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFG 114
+ ++ + L P VT D AL+ SVS DL + W +FG
Sbjct 442 -IRALSKEELQMP-VTRGDLELALKKIAKSVSAADLEKYEKWMVEFG 486
> ath:AT2G45500 ATP binding / nucleoside-triphosphatase/ nucleotide
binding
Length=487
Score = 35.4 bits (80), Expect = 0.044, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 1 SEGDFRYLARELEGFSGSDISVLVRDALFEPIR 33
S+GD + +E EG+SGSD+ L +A PIR
Sbjct 404 SDGDIDKIVKETEGYSGSDLQALCEEAAMMPIR 436
> cpv:cgd5_2010 nuclear VCP like protein with 2 AAA ATpase domains
; K14571 ribosome biogenesis ATPase
Length=695
Score = 34.7 bits (78), Expect = 0.073, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 24/34 (70%), Gaps = 0/34 (0%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRT 37
D R +++ +GFSG+D+S L+R+A + + + RT
Sbjct 599 DLRVISKNTQGFSGADLSQLIREATLKALDKLRT 632
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 10/24 (41%), Positives = 17/24 (70%), Gaps = 0/24 (0%)
Query 4 DFRYLARELEGFSGSDISVLVRDA 27
DFR ++R+ GF G+D+ L+ +A
Sbjct 280 DFREISRKTPGFVGADLKTLINEA 303
> pfa:PF07_0047 cell division cycle ATPase, putative
Length=1229
Score = 34.7 bits (78), Expect = 0.078, Method: Composition-based stats.
Identities = 29/119 (24%), Positives = 50/119 (42%), Gaps = 23/119 (19%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRC------RTATAFKQVMINGCAFFVPCHP 57
D +A+ EGFSG+DI+ L + A+ E I+ R +Q N +F +
Sbjct 1126 DIHDMAKRTEGFSGADITNLCQSAVNEAIKETIHLLNIRKKEQEEQRKKNKNSFKID-DT 1184
Query 58 GDSDPTARKMALMSVPADRLLPPEVTMEDFHSALRHTRPSVSPEDLRMHISWTEQFGLE 116
DP P ++ + F A ++ R S+ PED+ + + E+ L+
Sbjct 1185 DTYDPV----------------PTLSKKHFDLAFKNARISIQPEDVLKYEKFKEKLSLQ 1227
> tgo:TGME49_099170 ATPase, AAA family domain-containing protein
Length=831
Score = 34.3 bits (77), Expect = 0.090, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 4 DFRYLARELEGFSGSDISVLVRDALFEPIRRCRTATA 40
DFR LAR+ +G SG+D++ L +A R R A A
Sbjct 578 DFRLLARDSQGLSGADLACLCNEAALRASREGRAAVA 614
Lambda K H
0.323 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2027872200
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40