bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5651_orf2
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
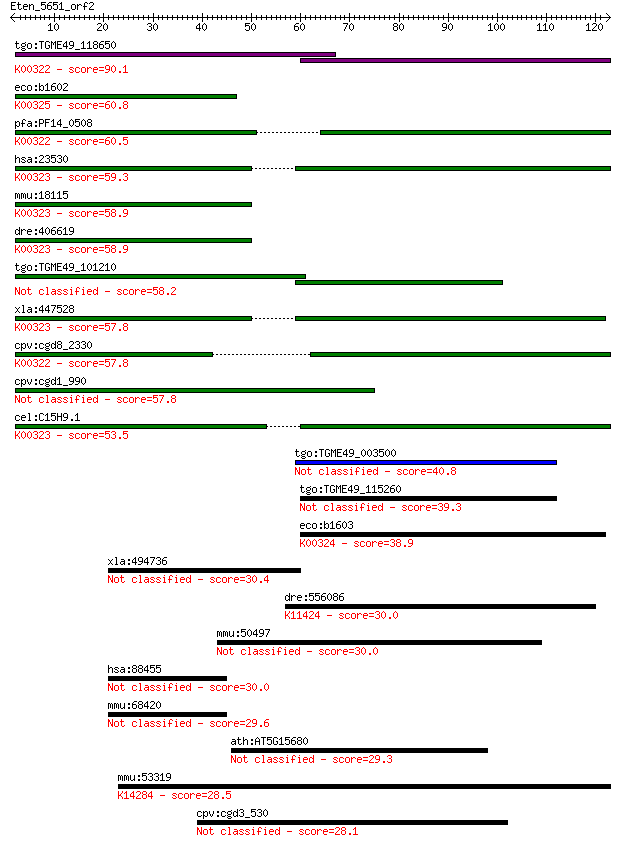
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 90.1 2e-18
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 60.8 1e-09
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 60.5 1e-09
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 59.3 3e-09
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 58.9 4e-09
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 58.9 4e-09
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 58.2 6e-09
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 57.8 8e-09
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 57.8 8e-09
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 57.8 9e-09
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 53.5 1e-07
tgo:TGME49_003500 alanine dehydrogenase, putative (EC:1.4.1.1) 40.8 0.001
tgo:TGME49_115260 alanine dehydrogenase, putative (EC:1.4.1.1) 39.3 0.003
eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydr... 38.9 0.004
xla:494736 ankrd13a; ankyrin repeat domain 13A 30.4 1.5
dre:556086 nsd1a; nuclear receptor binding SET domain protein ... 30.0 1.8
mmu:50497 Hspa14, 70kDa, HSP70L1, Hsp70-4, NST-1; heat shock p... 30.0 2.0
hsa:88455 ANKRD13A, ANKRD13, NY-REN-25; ankyrin repeat domain 13A 30.0 2.0
mmu:68420 Ankrd13a, 1100001D10Rik, ANKRD13, AU046136, MGC11850... 29.6 2.1
ath:AT5G15680 binding 29.3 3.6
mmu:53319 Nxf1, Mex67, Mvb1, Tap; nuclear RNA export factor 1 ... 28.5 6.0
cpv:cgd3_530 hypothetical protein 28.1 7.8
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 42/65 (64%), Positives = 53/65 (81%), Gaps = 0/65 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQMPPSAG 61
WKA+RV VLKRS+A GYA+I+NPLF L+NTRMLFGNAK+TT+A+FA ++ RA + SA
Sbjct 408 WKAKRVIVLKRSLAPGYAAIDNPLFFLDNTRMLFGNAKDTTTAIFACLSQRAAFVGKSAV 467
Query 62 FTDEE 66
F D E
Sbjct 468 FLDLE 472
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 30/63 (47%), Positives = 46/63 (73%), Gaps = 0/63 (0%)
Query 60 AGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVSRIPRDKVIISYVFPSINQQ 119
A F+D++Y + GAE++ D VI++S V+L+VS PS +L+ RIPR K++IS+VFP N
Sbjct 538 ANFSDDDYVKAGAEIMPNADTVISRSDVILKVSVPSEELIRRIPRGKILISHVFPGQNAP 597
Query 120 ALD 122
L+
Sbjct 598 LLE 600
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 26/45 (57%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVF 46
WKA+ V V KRSM GYA ++NPLF ENT MLFG+AK + A+
Sbjct 415 WKAQNVIVFKRSMNTGYAGVQNPLFFKENTHMLFGDAKASVDAIL 459
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 23/49 (46%), Positives = 38/49 (77%), Gaps = 0/49 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVN 50
WK+++V V KR++ GY++I+NPLF+ NT +LFG+AK+TT+ + +N
Sbjct 512 WKSKQVIVFKRTLNTGYSAIDNPLFYFSNTFLLFGDAKHTTNQILTILN 560
Score = 51.6 bits (122), Expect = 5e-07, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 40/59 (67%), Gaps = 1/59 (1%)
Query 64 DEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVSRIPRDKVIISYVFPSINQQALD 122
++EY + GAEV+S + ++ QS ++L+V P+ + + I + ++ISY++PSIN LD
Sbjct 686 NDEYTKYGAEVVS-RNVILQQSNIILKVDPPTVNFIEEIQNNTILISYLWPSINYHLLD 743
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 24/48 (50%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 49
WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V
Sbjct 1033 WKSKQVIVMKRSLGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQAKV 1080
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 18/70 (25%), Positives = 38/70 (54%), Gaps = 8/70 (11%)
Query 59 SAGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAP--SPDL----VSRIPRDKVIISYV 112
++ F+D+ YR GA++ + + S +V++V AP +P L + +IS++
Sbjct 98 ASKFSDDHYRVAGAQIQGAKEVL--ASDLVVKVRAPMVNPTLGVHEADLLKTSGTLISFI 155
Query 113 FPSINQQALD 122
+P+ N + L+
Sbjct 156 YPAQNPELLN 165
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 24/48 (50%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 49
WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V
Sbjct 1033 WKSKQVIVMKRSLGVGYAAVDNPIFYKPNTAMLLGDAKKTCDALQAKV 1080
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 24/48 (50%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 49
WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T A+ A+V
Sbjct 1029 WKSKQVVVMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDALSAKV 1076
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 25/59 (42%), Positives = 36/59 (61%), Gaps = 0/59 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQMPPSA 60
WKA++V VLKRSM GYA ++NPLF N+ ML G+AK + + + + + P A
Sbjct 400 WKAKKVIVLKRSMRVGYAGVDNPLFFYPNSEMLLGDAKASLQTLLTDMQDKLDAHPIEA 458
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 59 SAGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVS 100
SAGF DE+Y GA + + V+ S+VV++V+ D V+
Sbjct 570 SAGFLDEQYMESGALIADTAEEVVRLSRVVVKVTTFECDEVA 611
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 57.8 bits (138), Expect = 8e-09, Method: Composition-based stats.
Identities = 23/48 (47%), Positives = 37/48 (77%), Gaps = 0/48 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARV 49
WK+++V V+KRS+ GYA+++NP+F+ NT ML G+AK T ++ A+V
Sbjct 1033 WKSKQVIVMKRSLGVGYAAVDNPIFYKPNTSMLLGDAKKTCDSLQAKV 1080
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 8/69 (11%)
Query 59 SAGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAP--SPDL----VSRIPRDKVIISYV 112
++ F+D+ Y+ GA++ D + S +VL+V AP +P L ++S+V
Sbjct 98 ASKFSDDHYKEAGAKIQGTKDVL--ASDLVLKVRAPMLNPALGVHEADMFKPSSTLVSFV 155
Query 113 FPSINQQAL 121
+P+ N L
Sbjct 156 YPAQNPDLL 164
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 22/40 (55%), Positives = 32/40 (80%), Gaps = 0/40 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNT 41
W+A +V V KRS+ GYA+I+NPLF ++N M+FGNAK++
Sbjct 502 WRASKVIVSKRSLGKGYAAIDNPLFFMKNVEMIFGNAKDS 541
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 14/61 (22%), Positives = 37/61 (60%), Gaps = 0/61 (0%)
Query 62 FTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVSRIPRDKVIISYVFPSINQQAL 121
F D++Y +++ V+++S V+++V P+ + +S++ + ++SY++P+ N L
Sbjct 630 FDDQKYENASCTIMATRQDVVSRSDVIVKVQKPTDEEISQMKSGQTLVSYIWPAQNPSLL 689
Query 122 D 122
+
Sbjct 690 E 690
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 32/75 (42%), Positives = 42/75 (56%), Gaps = 9/75 (12%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQMPPSAG 61
WKA++V V KRS+A GYA IEN LF E TRML G+++ T V+ + AG
Sbjct 465 WKAKQVVVSKRSLAYGYACIENELFTCERTRMLLGDSRETLQQVYKILKG-------CAG 517
Query 62 FTDEEY--RRVGAEV 74
F +Y RV E+
Sbjct 518 FVPRQYIDSRVTEEI 532
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 36/51 (70%), Gaps = 0/51 (0%)
Query 2 WKARRVFVLKRSMAAGYASIENPLFHLENTRMLFGNAKNTTSAVFARVNAR 52
W +++V ++KR++ GYA+++NP+F ENT+ML G+AK + + V ++
Sbjct 989 WNSKQVIIVKRTLGTGYAAVDNPVFFNENTQMLLGDAKKMSEKLLEEVKSK 1039
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 38/63 (60%), Gaps = 2/63 (3%)
Query 60 AGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVSRIPRDKVIISYVFPSINQQ 119
AG+++EEY R GA+V G + + ++L+V P+ + VS++ +IS++ P NQ
Sbjct 71 AGYSNEEYVRSGADV--GKHNEVFNTDIMLKVRPPTENEVSKLKSGCTLISFIHPGQNQA 128
Query 120 ALD 122
LD
Sbjct 129 LLD 131
> tgo:TGME49_003500 alanine dehydrogenase, putative (EC:1.4.1.1)
Length=462
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 59 SAGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVSRIPRDKVIISY 111
++GFTDE Y GA ++ + V SQ++++V AP P S I +VI ++
Sbjct 69 ASGFTDESYTAAGATMVQTTEEVYKTSQMIVKVQAPQPQEYSFIQPGQVIFAF 121
> tgo:TGME49_115260 alanine dehydrogenase, putative (EC:1.4.1.1)
Length=390
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 32/52 (61%), Gaps = 0/52 (0%)
Query 60 AGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVSRIPRDKVIISY 111
A FTDE+Y + GAE+++ + + +S+++++V P PD + +++ Y
Sbjct 43 AHFTDEDYVQQGAEIVASAEELYGRSEMIVKVKEPQPDEWKLVKSGQILFCY 94
> eco:b1603 pntA, ECK1598, JW1595; pyridine nucleotide transhydrogenase,
alpha subunit (EC:1.6.1.2); K00324 NAD(P) transhydrogenase
subunit alpha [EC:1.6.1.2]
Length=510
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 39/62 (62%), Gaps = 2/62 (3%)
Query 60 AGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVSRIPRDKVIISYVFPSINQQ 119
A F D+ + + GAE++ G + QS+++L+V+AP D ++ + ++S+++P+ N +
Sbjct 43 ASFDDKAFVQAGAEIVEGNS--VWQSEIILKVNAPLDDEIALLNPGTTLVSFIWPAQNPE 100
Query 120 AL 121
+
Sbjct 101 LM 102
> xla:494736 ankrd13a; ankyrin repeat domain 13A
Length=593
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 19/39 (48%), Positives = 21/39 (53%), Gaps = 9/39 (23%)
Query 21 IENPLFHLENTRMLFGNAKNTTSAVFARVNARAEQMPPS 59
IE PLFH+ N R+ FGN NT S RAE P S
Sbjct 411 IEIPLFHVLNARITFGNV-NTCS--------RAEDSPAS 440
> dre:556086 nsd1a; nuclear receptor binding SET domain protein
1a; K11424 histone-lysine N-methyltransferase NSD1/2 [EC:2.1.1.43]
Length=2055
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 32/66 (48%), Gaps = 3/66 (4%)
Query 57 PPSAGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAP-SPDL--VSRIPRDKVIISYVF 113
P +G D E + S P AV QS+ LR S+P PD+ VS+IP + S
Sbjct 1114 PEESGVRDLEKQEEAGPPPSSPCAVSAQSEGELRTSSPIQPDVPSVSQIPVEGEQSSPSK 1173
Query 114 PSINQQ 119
P IN +
Sbjct 1174 PQINNE 1179
> mmu:50497 Hspa14, 70kDa, HSP70L1, Hsp70-4, NST-1; heat shock
protein 14
Length=509
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 35/66 (53%), Gaps = 3/66 (4%)
Query 43 SAVFARVNARAEQMPPSAGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDLVSRI 102
S +F + ++ GFT ++ +V VL G + I + Q +++ P+ DL++ I
Sbjct 305 SPLFNKCTEAIRELLRQTGFTADDINKV---VLCGGSSRIPKLQQLIKDLFPAVDLLNSI 361
Query 103 PRDKVI 108
P D+VI
Sbjct 362 PPDEVI 367
> hsa:88455 ANKRD13A, ANKRD13, NY-REN-25; ankyrin repeat domain
13A
Length=590
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 21 IENPLFHLENTRMLFGNAKNTTSA 44
IE PLFH+ N R+ FGN ++A
Sbjct 411 IEIPLFHVLNARITFGNVNGCSTA 434
> mmu:68420 Ankrd13a, 1100001D10Rik, ANKRD13, AU046136, MGC118502;
ankyrin repeat domain 13a
Length=588
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 21 IENPLFHLENTRMLFGNAKNTTSA 44
IE PLFH+ N R+ FGN ++A
Sbjct 411 IEIPLFHVLNARITFGNVNGCSTA 434
> ath:AT5G15680 binding
Length=2153
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 30/55 (54%), Gaps = 4/55 (7%)
Query 46 FARVNARAEQMPPSAGFTDEEYRRVGAEVLSGPDAVINQS---QVVLRVSAPSPD 97
F R +R +MPPS+G T ++ V VL G + ++ +V+ R++ PS D
Sbjct 1827 FQRSESRGYEMPPSSG-TLPKFEGVQPLVLKGLMSTVSHEFSIEVLSRITVPSCD 1880
> mmu:53319 Nxf1, Mex67, Mvb1, Tap; nuclear RNA export factor
1 homolog (S. cerevisiae); K14284 nuclear RNA export factor
1/2
Length=618
Score = 28.5 bits (62), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 30/109 (27%), Positives = 46/109 (42%), Gaps = 37/109 (33%)
Query 23 NPL-FHLENTRMLFGNAKNTTSAVFARVNARAEQMPPSAGFTDEEYRRVGAEVLSGPDAV 81
NP+ FH ENTR F TT++ VN + + D E RR+
Sbjct 147 NPIEFHYENTRAHFFVEDATTASALKGVNHKIQ---------DRENRRIS---------- 187
Query 82 INQSQVVLRVSAP--------SPDLVSRIPRDKVIISYVFPSINQQALD 122
+++ SAP P+ + ++ K+I+S + NQQALD
Sbjct 188 -----IIINASAPPYTVQNELKPEQIEQL---KLIMSKRYDG-NQQALD 227
> cpv:cgd3_530 hypothetical protein
Length=2285
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 18/63 (28%), Positives = 27/63 (42%), Gaps = 0/63 (0%)
Query 39 KNTTSAVFARVNARAEQMPPSAGFTDEEYRRVGAEVLSGPDAVINQSQVVLRVSAPSPDL 98
++ T +RV +R+ PS ++ E + + P A NQSQ V SP
Sbjct 78 ESKTLGELSRVKSRSADRSPSLPYSQEPSMELENSISKSPSAFPNQSQQYSPVYEQSPSK 137
Query 99 VSR 101
SR
Sbjct 138 NSR 140
Lambda K H
0.319 0.131 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2008132680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40