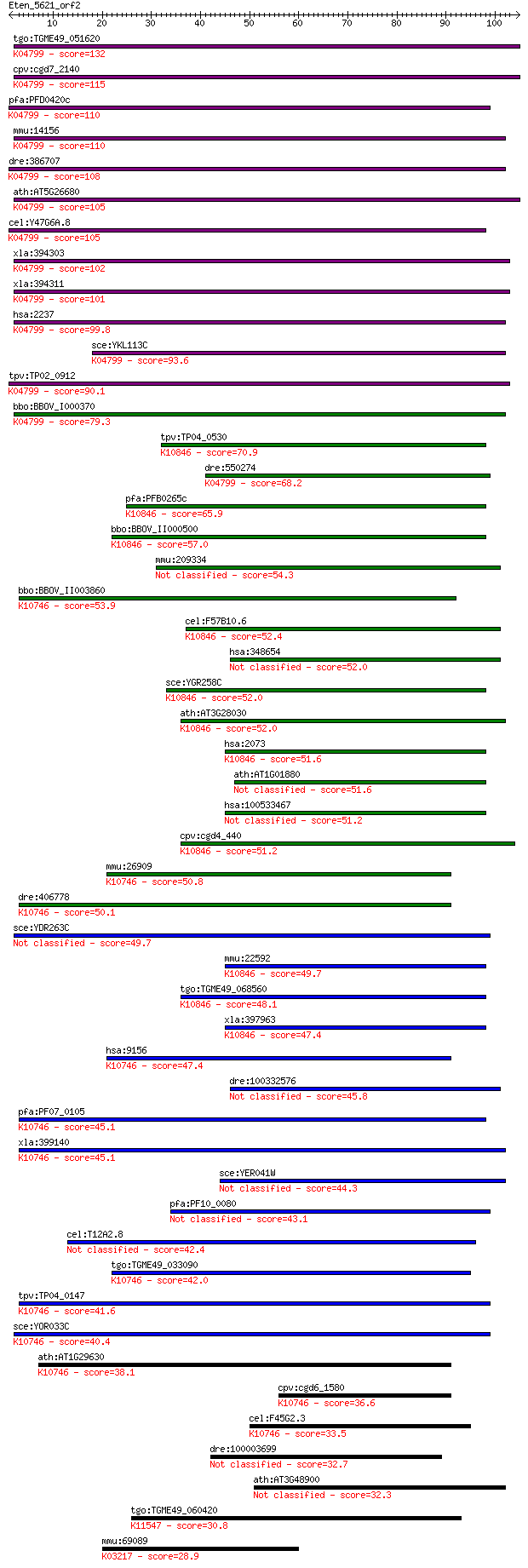
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5621_orf2
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_051620 flap endonuclease-1, putative ; K04799 flap ... 132 3e-31
cpv:cgd7_2140 flap endonuclease 1 ; K04799 flap endonuclease-1... 115 3e-26
pfa:PFD0420c FEN-1, PfFEN-1; flap endonuclease 1; K04799 flap ... 110 1e-24
mmu:14156 Fen1, AW538437; flap structure specific endonuclease... 110 2e-24
dre:386707 fen1, cb879; flap structure-specific endonuclease 1... 108 3e-24
ath:AT5G26680 endonuclease, putative; K04799 flap endonuclease... 105 4e-23
cel:Y47G6A.8 crn-1; Cell-death-Related Nuclease family member ... 105 4e-23
xla:394303 fen1-a, MGC196488, MGC196492, fen-1, fen1, mf1, rad... 102 2e-22
xla:394311 fen1-b, fen-1, mf1, rad2; flap structure-specific e... 101 7e-22
hsa:2237 FEN1, FEN-1, MF1, RAD2; flap structure-specific endon... 99.8 2e-21
sce:YKL113C RAD27, ERC11, FEN1, RTH1; 5' to 3' exonuclease, 5'... 93.6 2e-19
tpv:TP02_0912 flap endonuclease 1; K04799 flap endonuclease-1 ... 90.1 2e-18
bbo:BBOV_I000370 16.m00790; XPG N-terminal domain and XPG I-re... 79.3 2e-15
tpv:TP04_0530 DNA repair protein Rad2; K10846 DNA excision rep... 70.9 8e-13
dre:550274 im:7147072; zgc:110269; K04799 flap endonuclease-1 ... 68.2 6e-12
pfa:PFB0265c DNA repair endonuclease, putative; K10846 DNA exc... 65.9 3e-11
bbo:BBOV_II000500 18.m06025; Rad2 endonuclease; K10846 DNA exc... 57.0 1e-08
mmu:209334 Gen1, 5830483C08Rik, MGC115970; Gen homolog 1, endo... 54.3 1e-07
bbo:BBOV_II003860 18.m06322; XPG I-region family protein; K107... 53.9 1e-07
cel:F57B10.6 xpg-1; XPG (Xeroderma Pigmentosum group G) DNA re... 52.4 3e-07
hsa:348654 GEN1, DKFZp781F0986, FLJ40869, Gen; Gen homolog 1, ... 52.0 4e-07
sce:YGR258C RAD2; Rad2p; K10846 DNA excision repair protein ER... 52.0 4e-07
ath:AT3G28030 UVH3; UVH3 (ULTRAVIOLET HYPERSENSITIVE 3); DNA b... 52.0 5e-07
hsa:2073 ERCC5, COFS3, ERCM2, UVDR, XPG, XPGC; excision repair... 51.6 6e-07
ath:AT1G01880 DNA repair protein, putative 51.6 6e-07
hsa:100533467 BIVM-ERCC5; BIVM-ERCC5 readthrough 51.2 7e-07
cpv:cgd4_440 XPG, DNA excision repair protein, flap endonuclea... 51.2 7e-07
mmu:26909 Exo1, 5730442G03Rik, Msa; exonuclease 1; K10746 exon... 50.8 1e-06
dre:406778 exo1, fc16e04, wu:fc16e04; zgc:55521; K10746 exonuc... 50.1 2e-06
sce:YDR263C DIN7, DIN3; Din7p 49.7 2e-06
mmu:22592 Ercc5, MGC176031, Xpg; excision repair cross-complem... 49.7 2e-06
tgo:TGME49_068560 RAD2 endonuclease, putative ; K10846 DNA exc... 48.1 6e-06
xla:397963 ercc5, cofs3, ercm2, uvdr, xpg, xpgc; excision repa... 47.4 1e-05
hsa:9156 EXO1, HEX1, hExoI; exonuclease 1; K10746 exonuclease ... 47.4 1e-05
dre:100332576 Gen homolog 1, endonuclease-like 45.8 3e-05
pfa:PF07_0105 exonuclease I, putative; K10746 exonuclease 1 [E... 45.1 5e-05
xla:399140 exo1, MGC83785, exoi; exonuclease 1; K10746 exonucl... 45.1 6e-05
sce:YER041W YEN1; Holliday junction resolvase; localization is... 44.3 9e-05
pfa:PF10_0080 endonuclease, putative 43.1 2e-04
cel:T12A2.8 hypothetical protein 42.4 4e-04
tgo:TGME49_033090 exonuclease, putative (EC:3.4.21.72); K10746... 42.0 5e-04
tpv:TP04_0147 exonuclease I; K10746 exonuclease 1 [EC:3.1.-.-] 41.6 7e-04
sce:YOR033C EXO1, DHS1; 5'-3' exonuclease and flap-endonucleas... 40.4 0.002
ath:AT1G29630 DNA binding / catalytic/ nuclease; K10746 exonuc... 38.1 0.007
cpv:cgd6_1580 exonuclease i/din7p-like; xeroderma pigmentosum ... 36.6 0.018
cel:F45G2.3 hypothetical protein; K10746 exonuclease 1 [EC:3.1... 33.5 0.18
dre:100003699 hypothetical LOC100003699 32.7 0.26
ath:AT3G48900 DNA binding / catalytic/ chromatin binding / nuc... 32.3 0.34
tgo:TGME49_060420 HEC/Ndc80p family protein, putative ; K11547... 30.8 1.0
mmu:69089 Oxa1l, 1810020M02Rik, AI894287; oxidase assembly 1-l... 28.9 4.1
> tgo:TGME49_051620 flap endonuclease-1, putative ; K04799 flap
endonuclease-1 [EC:3.-.-.-]
Length=552
Score = 132 bits (331), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 66/103 (64%), Positives = 81/103 (78%), Gaps = 0/103 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+RRE RE+A AA A E G+ ELRKQ+ RSVRV+++ NEDVKRLL L+GLP+++AP E
Sbjct 101 KRRELRESAQEAAEKAREEGNVEELRKQIVRSVRVSKQHNEDVKRLLRLMGLPVVEAPCE 160
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDSSS 104
AEAQCA L + KVWA ATEDADALTFGA L+RNLTF + +S
Sbjct 161 AEAQCAELTKNRKVWATATEDADALTFGATRLIRNLTFGERAS 203
> cpv:cgd7_2140 flap endonuclease 1 ; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=490
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 54/103 (52%), Positives = 71/103 (68%), Gaps = 0/103 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R ERRE A A A E GD ++KQ R++ VT++Q EDVK+LL LG+P + AP E
Sbjct 101 KRDERREKALAELEKAQEIGDEELIKKQSVRTIHVTKKQVEDVKKLLGFLGMPCIDAPSE 160
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDSSS 104
AEAQCA L + G V+ TEDAD+LTFG PI ++ L FS+SS+
Sbjct 161 AEAQCAELCKDGLVYGVVTEDADSLTFGTPIQIKQLNFSESSN 203
> pfa:PFD0420c FEN-1, PfFEN-1; flap endonuclease 1; K04799 flap
endonuclease-1 [EC:3.-.-.-]
Length=672
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 51/98 (52%), Positives = 69/98 (70%), Gaps = 0/98 (0%)
Query 1 ERRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPG 60
E+R E+R+ A A E G+ E++KQ R+VRVTR+QNE+ K+LL L+G+P+++AP
Sbjct 100 EKRGEKRQKAEELLKKAKEEGNLEEIKKQSGRTVRVTRKQNEEAKKLLTLMGIPIIEAPC 159
Query 61 EAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT 98
EAE+QCA L + A ATEDADAL FG IL+RNL
Sbjct 160 EAESQCAFLTKYNLAHATATEDADALVFGTKILIRNLN 197
> mmu:14156 Fen1, AW538437; flap structure specific endonuclease
1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=380
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 55/100 (55%), Positives = 71/100 (71%), Gaps = 0/100 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R ERR A A EAG E+ K R V+VT++ N++ K LL L+G+P L AP E
Sbjct 99 KRSERRAEAEKQLQQAQEAGMEEEVEKFTKRLVKVTKQHNDECKHLLSLMGIPYLDAPSE 158
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSD 101
AEA CAALA+AGKV+AAATED D LTFG+P+L+R+LT S+
Sbjct 159 AEASCAALAKAGKVYAAATEDMDCLTFGSPVLMRHLTASE 198
> dre:386707 fen1, cb879; flap structure-specific endonuclease
1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=330
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 54/101 (53%), Positives = 71/101 (70%), Gaps = 0/101 (0%)
Query 1 ERRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPG 60
E+R ERR A A A EAG+ + K R V+VT++ NE+ K+LL L+G+P ++AP
Sbjct 98 EKRVERRAEAEKLLAQAQEAGEQENIDKFSKRLVKVTKQHNEECKKLLSLMGVPYIEAPC 157
Query 61 EAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSD 101
EAEA CAAL +AGKV+A ATED D LTFG +LLR+LT S+
Sbjct 158 EAEASCAALVKAGKVYATATEDMDGLTFGTTVLLRHLTASE 198
> ath:AT5G26680 endonuclease, putative; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=383
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 51/103 (49%), Positives = 72/103 (69%), Gaps = 2/103 (1%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R +R A A A EAG+ ++ K R+V+VT++ N+D KRLL L+G+P+++A E
Sbjct 100 KRYSKRADATADLTGAIEAGNKEDIEKYSKRTVKVTKQHNDDCKRLLRLMGVPVVEATSE 159
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDSSS 104
AEAQCAAL ++GKV+ A+ED D+LTFGAP LR+L D SS
Sbjct 160 AEAQCAALCKSGKVYGVASEDMDSLTFGAPKFLRHLM--DPSS 200
> cel:Y47G6A.8 crn-1; Cell-death-Related Nuclease family member
(crn-1); K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=382
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 52/97 (53%), Positives = 68/97 (70%), Gaps = 0/97 (0%)
Query 1 ERRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPG 60
E+R ERR A A A E GD E K R V+VT++QN++ KRLL L+G+P+++AP
Sbjct 98 EKRSERRAEAEKALTEAKEKGDVKEAEKFERRLVKVTKQQNDEAKRLLGLMGIPVVEAPC 157
Query 61 EAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
EAEAQCA L +AGKV+ TED DALTFG+ +LLR+
Sbjct 158 EAEAQCAHLVKAGKVFGTVTEDMDALTFGSTVLLRHF 194
> xla:394303 fen1-a, MGC196488, MGC196492, fen-1, fen1, mf1, rad2,
xFEN1a; flap structure-specific endonuclease 1; K04799
flap endonuclease-1 [EC:3.-.-.-]
Length=382
Score = 102 bits (255), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 56/101 (55%), Positives = 73/101 (72%), Gaps = 0/101 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R ERR A AA EAG+ + K R V+VT++ NE+ K+LL L+G+P + AP E
Sbjct 99 KRSERRAEAEKLLEAAEEAGEVENIEKFTKRLVKVTKQHNEECKKLLTLMGIPYVDAPCE 158
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDS 102
AEA CAAL +AGKV+AAATED DALTFG P+LLR+LT S++
Sbjct 159 AEATCAALVKAGKVYAAATEDMDALTFGTPVLLRHLTASEA 199
> xla:394311 fen1-b, fen-1, mf1, rad2; flap structure-specific
endonuclease 1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=382
Score = 101 bits (251), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 56/101 (55%), Positives = 73/101 (72%), Gaps = 0/101 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R ERR A AA EAG+ + K R V+VT++ NE+ K+LL L+G+P + AP E
Sbjct 99 KRSERRAEAEKLLEAAEEAGEVENIEKFNKRLVKVTKQHNEECKKLLSLMGIPYVDAPCE 158
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDS 102
AEA CAAL +AGKV+AAATED DALTFG P+LLR+LT S++
Sbjct 159 AEATCAALVKAGKVYAAATEDMDALTFGTPVLLRHLTASEA 199
> hsa:2237 FEN1, FEN-1, MF1, RAD2; flap structure-specific endonuclease
1; K04799 flap endonuclease-1 [EC:3.-.-.-]
Length=380
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 53/100 (53%), Positives = 69/100 (69%), Gaps = 0/100 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R ERR A A AG E+ K R V+VT++ N++ K LL L+G+P L AP E
Sbjct 99 KRSERRAEAEKQLQQAQAAGAEQEVEKFTKRLVKVTKQHNDECKHLLSLMGIPYLDAPSE 158
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSD 101
AEA CAAL +AGKV+AAATED D LTFG+P+L+R+LT S+
Sbjct 159 AEASCAALVKAGKVYAAATEDMDCLTFGSPVLMRHLTASE 198
> sce:YKL113C RAD27, ERC11, FEN1, RTH1; 5' to 3' exonuclease,
5' flap endonuclease, required for Okazaki fragment processing
and maturation as well as for long-patch base-excision repair;
member of the S. pombe RAD2/FEN1 family; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=382
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 46/84 (54%), Positives = 61/84 (72%), Gaps = 0/84 (0%)
Query 18 AEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWA 77
AEA E KQ R V+V++E NE+ ++LL L+G+P + AP EAEAQCA LA+ GKV+A
Sbjct 113 AEATTELEKMKQERRLVKVSKEHNEEAQKLLGLMGIPYIIAPTEAEAQCAELAKKGKVYA 172
Query 78 AATEDADALTFGAPILLRNLTFSD 101
AA+ED D L + P LLR+LTFS+
Sbjct 173 AASEDMDTLCYRTPFLLRHLTFSE 196
> tpv:TP02_0912 flap endonuclease 1; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=494
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 45/102 (44%), Positives = 64/102 (62%), Gaps = 0/102 (0%)
Query 1 ERRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPG 60
E+RR++RE A A+ A GD ++K + R+V+V++E NE K+LL L+G+P+++A
Sbjct 100 EKRRQQREEANASLKKAISEGDKESVKKLVGRTVKVSKEMNESAKKLLRLMGVPVIEALE 159
Query 61 EAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSDS 102
EAEAQCA L A+ED D L FG LLRN+ S S
Sbjct 160 EAEAQCAYLVTKNLCRFVASEDTDTLVFGGAFLLRNVASSSS 201
> bbo:BBOV_I000370 16.m00790; XPG N-terminal domain and XPG I-region
domain containing protein; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=672
Score = 79.3 bits (194), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 46/100 (46%), Positives = 68/100 (68%), Gaps = 0/100 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+R+ RE A ++ A E D +RK + R+VR+T+++NE K+LL L+G+P+++A E
Sbjct 101 KRKLLREEAESSLEKAIEEDDKEAIRKYVGRTVRITQKENESAKKLLRLVGVPVIEAAEE 160
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSD 101
AEAQCA L + G V A +EDADAL F +LL+NLT S+
Sbjct 161 AEAQCAYLCQRGFVTAVGSEDADALVFRCGVLLKNLTASN 200
> tpv:TP04_0530 DNA repair protein Rad2; K10846 DNA excision repair
protein ERCC-5
Length=835
Score = 70.9 bits (172), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 31/68 (45%), Positives = 46/68 (67%), Gaps = 2/68 (2%)
Query 32 RSVRVTREQ--NEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFG 89
+SV VT + +DV +LL G+P + AP EAE+QCA + R+GK +A ++D+D+L FG
Sbjct 517 QSVSVTHKNPYYDDVHKLLEHFGVPYIVAPSEAESQCAYMNRSGKCYAVISDDSDSLVFG 576
Query 90 APILLRNL 97
A LL+N
Sbjct 577 AKCLLKNF 584
> dre:550274 im:7147072; zgc:110269; K04799 flap endonuclease-1
[EC:3.-.-.-]
Length=350
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 32/58 (55%), Positives = 41/58 (70%), Gaps = 0/58 (0%)
Query 41 NEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT 98
N++ RLL L+G+P ++APGEAEA CA LA+ G V A A+ED D L FG +LLR L
Sbjct 110 NQECLRLLHLMGVPCIKAPGEAEALCAHLAKIGTVNAVASEDMDTLAFGGTVLLRQLN 167
> pfa:PFB0265c DNA repair endonuclease, putative; K10846 DNA excision
repair protein ERCC-5
Length=1516
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 41/73 (56%), Gaps = 0/73 (0%)
Query 25 ELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDAD 84
E +K ++ + E N+D+K LL G+P +Q+P EAEAQC+ L A ++D+D
Sbjct 1196 EYKKLKKNNIEINDEMNDDIKLLLNFFGIPYIQSPCEAEAQCSYLNNKNYCDAIISDDSD 1255
Query 85 ALTFGAPILLRNL 97
L F +++N
Sbjct 1256 VLVFSGKTVIKNF 1268
> bbo:BBOV_II000500 18.m06025; Rad2 endonuclease; K10846 DNA excision
repair protein ERCC-5
Length=1002
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 42/78 (53%), Gaps = 2/78 (2%)
Query 22 DAAELRKQLARSVRVTREQNE--DVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAA 79
D E+ +++R R +E V LL L G+P + AP EAEAQCA L G +
Sbjct 665 DFLEMENKISRMDRSQYRMDEWDKVPALLDLFGVPFIVAPSEAEAQCAHLNNTGLCFGVI 724
Query 80 TEDADALTFGAPILLRNL 97
++D+D L FGA + +N
Sbjct 725 SDDSDTLAFGAKRVFKNF 742
> mmu:209334 Gen1, 5830483C08Rik, MGC115970; Gen homolog 1, endonuclease
(Drosophila)
Length=908
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/74 (40%), Positives = 40/74 (54%), Gaps = 4/74 (5%)
Query 31 ARSVRVTREQNEDVKR----LLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADAL 86
+RS + R + V R +L LG+P +QA GEAEA CA L +G V T D DA
Sbjct 100 SRSQKTGRSHFKSVLRECLEMLECLGMPWVQAAGEAEAMCAYLNASGHVDGCLTNDGDAF 159
Query 87 TFGAPILLRNLTFS 100
+GA + RN T +
Sbjct 160 LYGAQTVYRNFTMN 173
> bbo:BBOV_II003860 18.m06322; XPG I-region family protein; K10746
exonuclease 1 [EC:3.1.-.-]
Length=501
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 51/92 (55%), Gaps = 3/92 (3%)
Query 3 RRERREAAAAAAAAAAEAGDAA---ELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAP 59
RRERR+ A A A E A E+ ++ +++ +T E V + + + ++ AP
Sbjct 104 RRERRDKAREEALAMIEKNGGAINTEIMRKCMQAIHITPEVIARVMEICRAMNVRIVVAP 163
Query 60 GEAEAQCAALARAGKVWAAATEDADALTFGAP 91
EA+AQ + L R+G +AA +ED+D L +G P
Sbjct 164 YEADAQVSYLCRSGIAYAALSEDSDLLVYGCP 195
> cel:F57B10.6 xpg-1; XPG (Xeroderma Pigmentosum group G) DNA
repair gene homolog family member (xpg-1); K10846 DNA excision
repair protein ERCC-5
Length=829
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 38/64 (59%), Gaps = 1/64 (1%)
Query 37 TREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRN 96
T E D++ L G+P +++PGEAEAQC L R G V ++D+D FGA + R+
Sbjct 474 TPELYRDLQEFLTNAGIPWIESPGEAEAQCVELERLGLVDGVVSDDSDVWAFGAQHVYRH 533
Query 97 LTFS 100
+ FS
Sbjct 534 M-FS 536
> hsa:348654 GEN1, DKFZp781F0986, FLJ40869, Gen; Gen homolog 1,
endonuclease (Drosophila)
Length=908
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 24/55 (43%), Positives = 30/55 (54%), Gaps = 0/55 (0%)
Query 46 RLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFS 100
+L LG+P +QA GEAEA CA L G V T D D +GA + RN T +
Sbjct 119 HMLECLGIPWVQAAGEAEAMCAYLNAGGHVDGCLTNDGDTFLYGAQTVYRNFTMN 173
> sce:YGR258C RAD2; Rad2p; K10846 DNA excision repair protein
ERCC-5
Length=1031
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 37/65 (56%), Gaps = 0/65 (0%)
Query 33 SVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPI 92
S VT + ++V+ LL G+P + AP EAEAQCA L + V T+D+D FG
Sbjct 764 SDEVTMDMIKEVQELLSRFGIPYITAPMEAEAQCAELLQLNLVDGIITDDSDVFLFGGTK 823
Query 93 LLRNL 97
+ +N+
Sbjct 824 IYKNM 828
> ath:AT3G28030 UVH3; UVH3 (ULTRAVIOLET HYPERSENSITIVE 3); DNA
binding / catalytic/ endonuclease/ nuclease/ single-stranded
DNA binding; K10846 DNA excision repair protein ERCC-5
Length=1479
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 36 VTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLR 95
V+ E + + LL + G+P + AP EAEAQCA + ++ V T+D+D FGA + +
Sbjct 921 VSSEMFAECQELLQIFGIPYIIAPMEAEAQCAFMEQSNLVDGIVTDDSDVFLFGARSVYK 980
Query 96 NLTFSD 101
N+ F D
Sbjct 981 NI-FDD 985
> hsa:2073 ERCC5, COFS3, ERCM2, UVDR, XPG, XPGC; excision repair
cross-complementing rodent repair deficiency, complementation
group 5; K10846 DNA excision repair protein ERCC-5
Length=1186
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 45 KRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
+ LL L G+P +QAP EAEAQCA L + T+D+D FGA + RN
Sbjct 773 QELLRLFGIPYIQAPMEAEAQCAILDLTDQTSGTITDDSDIWLFGARHVYRNF 825
> ath:AT1G01880 DNA repair protein, putative
Length=570
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 26/51 (50%), Positives = 35/51 (68%), Gaps = 0/51 (0%)
Query 47 LLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
LL LLG+P+L+A GEAEA CA L G V A T D+DA FGA +++++
Sbjct 126 LLELLGIPVLKANGEAEALCAQLNSQGFVDACITPDSDAFLFGAMCVIKDI 176
> hsa:100533467 BIVM-ERCC5; BIVM-ERCC5 readthrough
Length=1640
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 24/53 (45%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 45 KRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
+ LL L G+P +QAP EAEAQCA L + T+D+D FGA + RN
Sbjct 1227 QELLRLFGIPYIQAPMEAEAQCAILDLTDQTSGTITDDSDIWLFGARHVYRNF 1279
> cpv:cgd4_440 XPG, DNA excision repair protein, flap endonuclease
; K10846 DNA excision repair protein ERCC-5
Length=1147
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 39/68 (57%), Gaps = 1/68 (1%)
Query 36 VTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLR 95
+T+E + LL LG+P + +PGEAEAQ + L + ++D+D L FGA + R
Sbjct 875 ITKEMQYQICLLLKALGIPWIDSPGEAEAQASILTQLNICHGVLSDDSDCLIFGAKKIFR 934
Query 96 NLTFSDSS 103
N FS +S
Sbjct 935 NF-FSGNS 941
> mmu:26909 Exo1, 5730442G03Rik, Msa; exonuclease 1; K10746 exonuclease
1 [EC:3.1.-.-]
Length=837
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/70 (42%), Positives = 38/70 (54%), Gaps = 0/70 (0%)
Query 21 GDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAAT 80
G +E R ARS+ +T V + LG+ L AP EA+AQ A L +AG V A T
Sbjct 110 GKVSEARDCFARSINITHAMAHKVIKAARALGVDCLVAPYEADAQLAYLNKAGIVQAVIT 169
Query 81 EDADALTFGA 90
ED+D L FG
Sbjct 170 EDSDLLAFGC 179
> dre:406778 exo1, fc16e04, wu:fc16e04; zgc:55521; K10746 exonuclease
1 [EC:3.1.-.-]
Length=806
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/88 (37%), Positives = 42/88 (47%), Gaps = 0/88 (0%)
Query 3 RRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEA 62
RRERR+A G E R+ RSV +T DV R G+ + AP EA
Sbjct 92 RRERRQANLQKGKQLLREGKITEARECFTRSVNITPSMAHDVIRAARTRGVDCVVAPYEA 151
Query 63 EAQCAALARAGKVWAAATEDADALTFGA 90
+AQ A L ++ A TED+D L FG
Sbjct 152 DAQLAFLNKSDIAQAVITEDSDLLAFGC 179
> sce:YDR263C DIN7, DIN3; Din7p
Length=430
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 44/97 (45%), Gaps = 0/97 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
RRR++R A AG+ + +SV +T E + + L +P + AP E
Sbjct 91 RRRKKRLENEMIAKKLWSAGNRYNAMEYFQKSVDITPEMAKCIIDYCKLHSIPYIVAPFE 150
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT 98
A+ Q L + G + +ED+D L FG L+ L
Sbjct 151 ADPQMVYLEKMGLIQGIISEDSDLLVFGCKTLITKLN 187
> mmu:22592 Ercc5, MGC176031, Xpg; excision repair cross-complementing
rodent repair deficiency, complementation group 5;
K10846 DNA excision repair protein ERCC-5
Length=1170
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/53 (43%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 45 KRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
+ LL L G+P +QAP EAEAQCA L + T+D+D FGA + +N
Sbjct 772 QELLRLFGVPYIQAPMEAEAQCAMLDLTDQTSGTITDDSDIWLFGARHVYKNF 824
> tgo:TGME49_068560 RAD2 endonuclease, putative ; K10846 DNA excision
repair protein ERCC-5
Length=2004
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 25/62 (40%), Positives = 34/62 (54%), Gaps = 0/62 (0%)
Query 36 VTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLR 95
+T + V LL G+P + APGEAEA A L + A ++D+DAL FGA + R
Sbjct 1421 ITERMKDQVIALLRAFGVPFITAPGEAEATAAYLTKQNLADAVISDDSDALVFGAREIYR 1480
Query 96 NL 97
N
Sbjct 1481 NF 1482
> xla:397963 ercc5, cofs3, ercm2, uvdr, xpg, xpgc; excision repair
cross-complementing rodent repair deficiency, complementation
group 5; K10846 DNA excision repair protein ERCC-5
Length=1197
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/53 (41%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 45 KRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
+ LL L G+P + AP EAEAQCA L + T+D+D FGA + +N
Sbjct 807 QELLQLFGIPYIVAPMEAEAQCAILDLTDQTSGTITDDSDIWLFGARHVYKNF 859
> hsa:9156 EXO1, HEX1, hExoI; exonuclease 1; K10746 exonuclease
1 [EC:3.1.-.-]
Length=803
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 37/70 (52%), Gaps = 0/70 (0%)
Query 21 GDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAAT 80
G +E R+ RS+ +T V + G+ L AP EA+AQ A L +AG V A T
Sbjct 110 GKVSEARECFTRSINITHAMAHKVIKAARSQGVDCLVAPYEADAQLAYLNKAGIVQAIIT 169
Query 81 EDADALTFGA 90
ED+D L FG
Sbjct 170 EDSDLLAFGC 179
> dre:100332576 Gen homolog 1, endonuclease-like
Length=179
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 30/55 (54%), Gaps = 0/55 (0%)
Query 46 RLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFS 100
++L LG+P + A GEAEA CA L G V T D DA +GA + RN +
Sbjct 58 QMLDCLGVPWVTAAGEAEAMCAFLDSQGLVDGCITNDGDAFLYGARTVYRNFNMT 112
> pfa:PF07_0105 exonuclease I, putative; K10746 exonuclease 1
[EC:3.1.-.-]
Length=1347
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 47/99 (47%), Gaps = 4/99 (4%)
Query 3 RRERREAAAA----AAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQA 58
R+ RRE A + + K+ +++ V++E + VK + + +
Sbjct 92 RKNRREKAKMELQEIISKVKNPRTNEMVLKKCIQAISVSKEIIDSVKEFCRKKNIDYIIS 151
Query 59 PGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNL 97
P EA+AQ + L R G + A +ED+D L +G P +L L
Sbjct 152 PYEADAQLSYLCRMGFISCAISEDSDLLVYGCPRVLYKL 190
> xla:399140 exo1, MGC83785, exoi; exonuclease 1; K10746 exonuclease
1 [EC:3.1.-.-]
Length=734
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 48/100 (48%), Gaps = 1/100 (1%)
Query 3 RRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGEA 62
RRE+R+ G AE R+ +RSV +T +V + G+ + AP EA
Sbjct 92 RREKRQTNLQKGKQLLREGKLAEARECFSRSVNITSSMAHEVIKAARSEGVDYIVAPYEA 151
Query 63 EAQCAALARAGKVWAAATEDADALTFGA-PILLRNLTFSD 101
++Q A L + A TED+D L FG +LL+ F +
Sbjct 152 DSQLAYLNKNDFAEAIITEDSDLLAFGCKKVLLKMDKFGN 191
> sce:YER041W YEN1; Holliday junction resolvase; localization
is cell-cycle dependent and regulated by Cdc28p phosphorylation;
homolog of human GEN1 and has similarity to S. cerevisiae
endonuclease Rth1p
Length=759
Score = 44.3 bits (103), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 23/59 (38%), Positives = 34/59 (57%), Gaps = 1/59 (1%)
Query 44 VKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT-FSD 101
V++LL L+ + + A GE EAQC L +G V + D+D L FG +L+N + F D
Sbjct 176 VRKLLDLMNISYVIACGEGEAQCVWLQVSGAVDFILSNDSDTLVFGGEKILKNYSKFYD 234
> pfa:PF10_0080 endonuclease, putative
Length=388
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Query 34 VRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPIL 93
+++ + D+ LLL +P+ +AE +CA + ++D DAL FGAP L
Sbjct 218 IKINSKTANDIYNYLLLENIPIFITKNDAEKECAIQCSHERD-IVVSDDTDALAFGAPNL 276
Query 94 LRNLT 98
+R +T
Sbjct 277 IRFIT 281
> cel:T12A2.8 hypothetical protein
Length=434
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 30/88 (34%), Positives = 42/88 (47%), Gaps = 5/88 (5%)
Query 13 AAAAAAEAGDAAELRKQLARSVRVTREQN--EDVKR---LLLLLGLPLLQAPGEAEAQCA 67
A+++A E+ D E + RS + N + V + LL LG+ ++ APG+ EAQCA
Sbjct 80 ASSSAHESKDQNEFVPRKRRSFGDSPFTNLVDHVYKTNALLTELGIKVIIAPGDGEAQCA 139
Query 68 ALARAGKVWAAATEDADALTFGAPILLR 95
L G T D D FG L R
Sbjct 140 RLEDLGVTSGCITTDFDYFLFGGKNLYR 167
> tgo:TGME49_033090 exonuclease, putative (EC:3.4.21.72); K10746
exonuclease 1 [EC:3.1.-.-]
Length=951
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 43/78 (55%), Gaps = 5/78 (6%)
Query 22 DAAELRKQLARSVRVTREQNEDVKRLLLL-----LGLPLLQAPGEAEAQCAALARAGKVW 76
+A EL K+ + R R+ D + ++ LG+ + AP EA+AQ A LAR GK+
Sbjct 124 EARELLKKYEEARRAGRKPPGDTRVDTVISACRSLGVAFVVAPYEADAQLAFLARTGKIA 183
Query 77 AAATEDADALTFGAPILL 94
AA +ED+D L G +L
Sbjct 184 AAVSEDSDLLAHGCQQVL 201
> tpv:TP04_0147 exonuclease I; K10746 exonuclease 1 [EC:3.1.-.-]
Length=550
Score = 41.6 bits (96), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 49/99 (49%), Gaps = 3/99 (3%)
Query 3 RRERREAAAAAAAAAAEAGDA---AELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAP 59
RRERR A A A E+ ++ +++++T E V + + + ++ +P
Sbjct 103 RRERRNKARAEALEMIRKNKGKINTEIMRKCMQAIQITPEIVHRVITICKKVNVSVVVSP 162
Query 60 GEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT 98
EA+AQ + L R G A +ED+D + +G P ++ L
Sbjct 163 YEADAQISYLCRTGIADFAISEDSDLIVYGCPKIIFKLN 201
> sce:YOR033C EXO1, DHS1; 5'-3' exonuclease and flap-endonuclease
involved in recombination, double-strand break repair and
DNA mismatch repair; member of the Rad2p nuclease family,
with conserved N and I nuclease domains (EC:3.1.-.-); K10746
exonuclease 1 [EC:3.1.-.-]
Length=702
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 43/97 (44%), Gaps = 0/97 (0%)
Query 2 RRRERREAAAAAAAAAAEAGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAPGE 61
+RR++R+ A A G+ + V +T E + + L G+ + AP E
Sbjct 91 KRRDKRKENKAIAERLWACGEKKNAMDYFQKCVDITPEMAKCIICYCKLNGIRYIVAPFE 150
Query 62 AEAQCAALARAGKVWAAATEDADALTFGAPILLRNLT 98
A++Q L + V +ED+D L FG L+ L
Sbjct 151 ADSQMVYLEQKNIVQGIISEDSDLLVFGCRRLITKLN 187
> ath:AT1G29630 DNA binding / catalytic/ nuclease; K10746 exonuclease
1 [EC:3.1.-.-]
Length=735
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 41/85 (48%), Gaps = 12/85 (14%)
Query 7 REAAAAAAAAAAEAGDAAELRKQLARS-VRVTREQNEDVKRLLLLLGLPLLQAPGEAEAQ 65
EA ++AA A ++ +A ++V R++N D + AP EA+AQ
Sbjct 106 HEANGNSSAAYECYSKAVDISPSIAHELIQVLRQENVD-----------YVVAPYEADAQ 154
Query 66 CAALARAGKVWAAATEDADALTFGA 90
A LA +V A TED+D + FG
Sbjct 155 MAFLAITKQVDAIITEDSDLIPFGC 179
> cpv:cgd6_1580 exonuclease i/din7p-like; xeroderma pigmentosum
G N-region plus xeroderma pigmentosum G I-region plus HhH2
domain ; K10746 exonuclease 1 [EC:3.1.-.-]
Length=482
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 56 LQAPGEAEAQCAALARAGKVWAAATEDADALTFGA 90
+ AP EA+AQ + L+R + A TED+D L FG+
Sbjct 149 IVAPYEADAQLSYLSRIKYIDAVITEDSDMLVFGS 183
> cel:F45G2.3 hypothetical protein; K10746 exonuclease 1 [EC:3.1.-.-]
Length=639
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 26/45 (57%), Gaps = 0/45 (0%)
Query 50 LLGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILL 94
+ + ++ AP EA+AQ A L + V A TED+D + FG ++
Sbjct 140 MTNVDIVVAPYEADAQLAYLVQEKLVDAVITEDSDLIVFGCEMIY 184
> dre:100003699 hypothetical LOC100003699
Length=469
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 27/49 (55%), Gaps = 5/49 (10%)
Query 42 EDVKRLLLLLGLPLLQAPGEAEAQCAALARAGKVWAAA--TEDADALTF 88
E K++L LG+PL+Q EA+ + A+LA K W T D+D F
Sbjct 120 EVFKQVLFELGVPLVQCISEADFEIASLA---KQWGCPVLTNDSDFFIF 165
> ath:AT3G48900 DNA binding / catalytic/ chromatin binding / nuclease
Length=600
Score = 32.3 bits (72), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 51 LGLPLLQAPGEAEAQCAALARAGKVWAAATEDADALTFGAPILLRNLTFSD 101
LG+ L EAEAQCA L A + D+D FGA + R + +
Sbjct 132 LGILCLDGIEEAEAQCALLNSESLCDACFSFDSDIFLFGAKTVYREICLGE 182
> tgo:TGME49_060420 HEC/Ndc80p family protein, putative ; K11547
kinetochore protein NDC80
Length=663
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 39/90 (43%), Gaps = 23/90 (25%)
Query 26 LRKQLARSVRVTREQ-NEDVKRLLLLLGLPL------LQAPGEAEAQ----------CAA 68
LR+ SV+VT E NE+V RL LG PL +QAP A C
Sbjct 183 LRRACDDSVQVTNENANEEVPRLFKELGYPLTIAKSSMQAPNSAHQWPLHLHALSWLCEL 242
Query 69 LARAGKVWA------AATEDADALTFGAPI 92
L +V++ A E DAL+ GA +
Sbjct 243 LIYESEVFSRDPLLNPAIEKKDALSAGAVV 272
> mmu:69089 Oxa1l, 1810020M02Rik, AI894287; oxidase assembly 1-like;
K03217 preprotein translocase subunit YidC
Length=433
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 3/40 (7%)
Query 20 AGDAAELRKQLARSVRVTREQNEDVKRLLLLLGLPLLQAP 59
AGD AE K ++ +TR Q + +LL L LPL QAP
Sbjct 184 AGDQAEFYKA---TIEMTRYQKKHDIKLLRPLILPLTQAP 220
Lambda K H
0.315 0.125 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022291452
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40