bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5606_orf3
Length=81
Score E
Sequences producing significant alignments: (Bits) Value
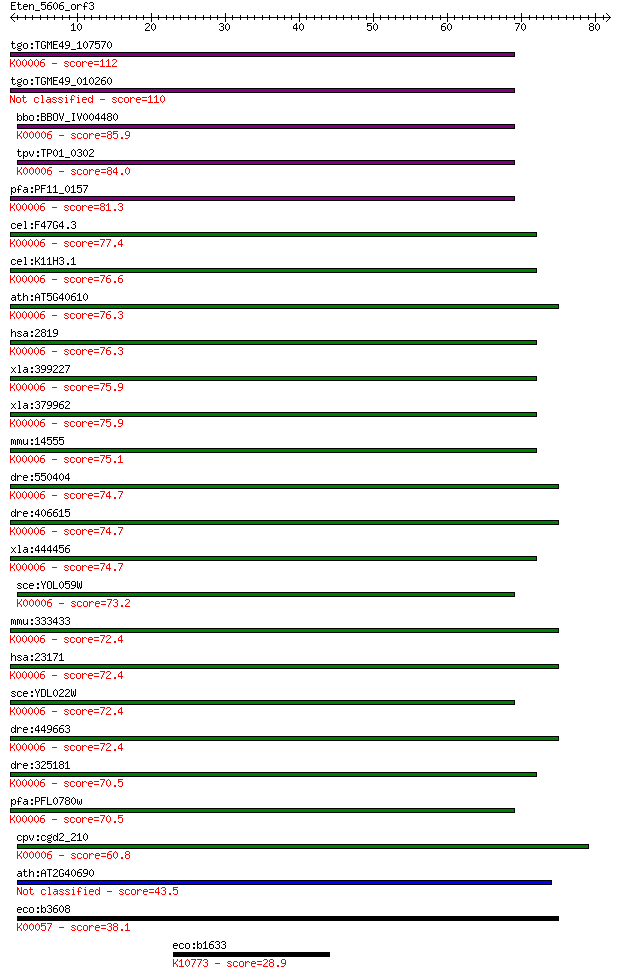
tgo:TGME49_107570 glycerol-3-phosphate dehydrogenase, putative... 112 2e-25
tgo:TGME49_010260 glycerol-3-phosphate dehydrogenase, putative... 110 1e-24
bbo:BBOV_IV004480 23.m06005; glycerol-3-phosphate dehydrogenas... 85.9 3e-17
tpv:TP01_0302 NAD-dependent glycerol-3-phosphate dehydrogenase... 84.0 1e-16
pfa:PF11_0157 glycerol-3-phosphate dehydrogenase, putative; K0... 81.3 8e-16
cel:F47G4.3 gpdh-1; Glycerol-3-Phosphate DeHydrogenase family ... 77.4 1e-14
cel:K11H3.1 gpdh-2; Glycerol-3-Phosphate DeHydrogenase family ... 76.6 2e-14
ath:AT5G40610 glycerol-3-phosphate dehydrogenase (NAD+) / GPDH... 76.3 2e-14
hsa:2819 GPD1, FLJ26652; glycerol-3-phosphate dehydrogenase 1 ... 76.3 2e-14
xla:399227 gpd1; glycerol-3-phosphate dehydrogenase 1 (soluble... 75.9 3e-14
xla:379962 gpd1l, MGC130781, MGC52863; glycerol-3-phosphate de... 75.9 3e-14
mmu:14555 Gpd1, AI747587, Gdc-1, Gdc1, KIAA4010, mKIAA4010; gl... 75.1 5e-14
dre:550404 gpd1a, zgc:112197; glycerol-3-phosphate dehydrogena... 74.7 7e-14
dre:406615 gpd1, wu:fc30a07, zgc:63859; glycerol-3-phosphate d... 74.7 7e-14
xla:444456 MGC83663 protein; K00006 glycerol-3-phosphate dehyd... 74.7 7e-14
sce:YOL059W GPD2, GPD3; Gpd2p (EC:1.1.1.8); K00006 glycerol-3-... 73.2 2e-13
mmu:333433 Gpd1l, 2210409H23Rik, D9Ertd660e; glycerol-3-phosph... 72.4 3e-13
hsa:23171 GPD1L, GPD1-L, KIAA0089; glycerol-3-phosphate dehydr... 72.4 3e-13
sce:YDL022W GPD1, DAR1, HOR1, OSG1, OSR5; Gpd1p; K00006 glycer... 72.4 3e-13
dre:449663 gpd1l, wu:fi13g03, wu:fi45b08, zgc:92580; glycerol-... 72.4 4e-13
dre:325181 gpd1b, Gpd1, gpd1h, gpd1l, wu:fc58b05, zgc:66051, z... 70.5 1e-12
pfa:PFL0780w glycerol-3-phosphate dehydrogenase, putative (EC:... 70.5 1e-12
cpv:cgd2_210 glycerol-3-phosphate dehydrogenase (EC:1.1.1.8); ... 60.8 1e-09
ath:AT2G40690 GLY1; GLY1; glycerol-3-phosphate dehydrogenase (... 43.5 2e-04
eco:b3608 gpsA, ECK3598, JW3583; glycerol-3-phosphate dehydrog... 38.1 0.007
eco:b1633 nth, ECK1629, JW1625; DNA glycosylase and apyrimidin... 28.9 4.4
> tgo:TGME49_107570 glycerol-3-phosphate dehydrogenase, putative
(EC:1.1.1.8); K00006 glycerol-3-phosphate dehydrogenase (NAD+)
[EC:1.1.1.8]
Length=401
Score = 112 bits (281), Expect = 2e-25, Method: Composition-based stats.
Identities = 50/68 (73%), Positives = 61/68 (89%), Gaps = 0/68 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGA+KNVVALA GFC+G+G GTNTK+AI+RLGVEE+K F M FF +++A+T +DSAGYAD
Sbjct 224 CGAVKNVVALAAGFCDGLGLGTNTKSAIIRLGVEEMKQFGMIFFDNVIAETFFDSAGYAD 283
Query 61 VITTVFGG 68
VITTVFGG
Sbjct 284 VITTVFGG 291
> tgo:TGME49_010260 glycerol-3-phosphate dehydrogenase, putative
(EC:1.1.1.8)
Length=515
Score = 110 bits (275), Expect = 1e-24, Method: Composition-based stats.
Identities = 47/68 (69%), Positives = 59/68 (86%), Gaps = 0/68 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGA+KNV+A+A GFC+G+G GTN K AI+RLGVEE+K F M FF +++A+T +DSAGYAD
Sbjct 218 CGAVKNVIAIAAGFCDGLGLGTNAKTAIIRLGVEEMKQFAMIFFDNIMAETFFDSAGYAD 277
Query 61 VITTVFGG 68
VITTVFGG
Sbjct 278 VITTVFGG 285
> bbo:BBOV_IV004480 23.m06005; glycerol-3-phosphate dehydrogenase
(EC:1.1.1.8); K00006 glycerol-3-phosphate dehydrogenase
(NAD+) [EC:1.1.1.8]
Length=354
Score = 85.9 bits (211), Expect = 3e-17, Method: Composition-based stats.
Identities = 38/67 (56%), Positives = 51/67 (76%), Gaps = 0/67 (0%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYADV 61
GA+KN VA+A GFC+G+G G+NTKAAI+R+G+ EI F FF D+ D +++SAG AD+
Sbjct 204 GAIKNAVAIAAGFCDGLGLGSNTKAAIMRIGLNEIYRFACKFFKDINTDVVFESAGVADL 263
Query 62 ITTVFGG 68
ITT GG
Sbjct 264 ITTCIGG 270
> tpv:TP01_0302 NAD-dependent glycerol-3-phosphate dehydrogenase;
K00006 glycerol-3-phosphate dehydrogenase (NAD+) [EC:1.1.1.8]
Length=354
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 37/67 (55%), Positives = 51/67 (76%), Gaps = 0/67 (0%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYADV 61
GA+KNVVAL+ GFC+G+G G+NTKAA++R+G+ EI F FF + D +++SAG AD+
Sbjct 203 GAIKNVVALSAGFCDGLGLGSNTKAAVMRIGLVEIHKFAKLFFPNTTEDVVFESAGVADL 262
Query 62 ITTVFGG 68
ITT GG
Sbjct 263 ITTCIGG 269
> pfa:PF11_0157 glycerol-3-phosphate dehydrogenase, putative;
K00006 glycerol-3-phosphate dehydrogenase (NAD+) [EC:1.1.1.8]
Length=394
Score = 81.3 bits (199), Expect = 8e-16, Method: Composition-based stats.
Identities = 37/68 (54%), Positives = 49/68 (72%), Gaps = 0/68 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGALKNV+AL GF +G+ NTK+AI+R+G+EE+K F FF ++L +T DS G AD
Sbjct 240 CGALKNVIALGVGFIDGLTASYNTKSAIIRIGLEEMKKFAKMFFPNVLDETFLDSCGLAD 299
Query 61 VITTVFGG 68
+ITT GG
Sbjct 300 LITTCLGG 307
> cel:F47G4.3 gpdh-1; Glycerol-3-Phosphate DeHydrogenase family
member (gpdh-1); K00006 glycerol-3-phosphate dehydrogenase
(NAD+) [EC:1.1.1.8]
Length=374
Score = 77.4 bits (189), Expect = 1e-14, Method: Composition-based stats.
Identities = 35/71 (49%), Positives = 48/71 (67%), Gaps = 0/71 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGALKN+VA A GF +G+G N K+AI+RLG+ E K F+ F+ + T ++S G AD
Sbjct 226 CGALKNIVACAAGFADGLGWAYNVKSAIIRLGLLETKKFVEHFYPSSVGHTYFESCGVAD 285
Query 61 VITTVFGGNCR 71
+ITT +GG R
Sbjct 286 LITTCYGGRNR 296
> cel:K11H3.1 gpdh-2; Glycerol-3-Phosphate DeHydrogenase family
member (gpdh-2); K00006 glycerol-3-phosphate dehydrogenase
(NAD+) [EC:1.1.1.8]
Length=371
Score = 76.6 bits (187), Expect = 2e-14, Method: Composition-based stats.
Identities = 36/71 (50%), Positives = 48/71 (67%), Gaps = 0/71 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGALKNVVA A GF +G+G G NTKAA++RLG+ E F+ ++ T ++S G AD
Sbjct 223 CGALKNVVACAAGFTDGLGYGDNTKAAVIRLGLMETTKFVEHYYPGSNLQTFFESCGIAD 282
Query 61 VITTVFGGNCR 71
+ITT +GG R
Sbjct 283 LITTCYGGRNR 293
> ath:AT5G40610 glycerol-3-phosphate dehydrogenase (NAD+) / GPDH;
K00006 glycerol-3-phosphate dehydrogenase (NAD+) [EC:1.1.1.8]
Length=400
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/74 (51%), Positives = 48/74 (64%), Gaps = 2/74 (2%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CG LKNVVA+A GF +G+ G NTKAAI+R+G+ E+K F + T ++S G AD
Sbjct 253 CGTLKNVVAIAAGFVDGLEMGNNTKAAIMRIGLREMKALSKLLFPSVKDSTFFESCGVAD 312
Query 61 VITTVFGGNCRARR 74
VITT GG R RR
Sbjct 313 VITTCLGG--RNRR 324
> hsa:2819 GPD1, FLJ26652; glycerol-3-phosphate dehydrogenase
1 (soluble) (EC:1.1.1.8); K00006 glycerol-3-phosphate dehydrogenase
(NAD+) [EC:1.1.1.8]
Length=349
Score = 76.3 bits (186), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/72 (52%), Positives = 50/72 (69%), Gaps = 1/72 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTF-FGDLLADTLYDSAGYA 59
CGALKNVVA+ GFC+G+G G NTKAA++RLG+ E+ F F G + + T +S G A
Sbjct 200 CGALKNVVAVGAGFCDGLGFGDNTKAAVIRLGLMEMIAFAKLFCSGPVSSATFLESCGVA 259
Query 60 DVITTVFGGNCR 71
D+ITT +GG R
Sbjct 260 DLITTCYGGRNR 271
> xla:399227 gpd1; glycerol-3-phosphate dehydrogenase 1 (soluble)
(EC:1.1.1.8); K00006 glycerol-3-phosphate dehydrogenase
(NAD+) [EC:1.1.1.8]
Length=348
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 37/72 (51%), Positives = 50/72 (69%), Gaps = 1/72 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTF-FGDLLADTLYDSAGYA 59
CGALKN+VA+ GFC+G+G G NTKAA++RLG+ E+ F F G + + T +S G A
Sbjct 199 CGALKNIVAVGAGFCDGLGFGDNTKAAVIRLGLMEMIAFSKLFCTGSVTSATFLESCGVA 258
Query 60 DVITTVFGGNCR 71
D+ITT +GG R
Sbjct 259 DLITTCYGGRNR 270
> xla:379962 gpd1l, MGC130781, MGC52863; glycerol-3-phosphate
dehydrogenase 1-like (EC:1.1.1.8); K00006 glycerol-3-phosphate
dehydrogenase (NAD+) [EC:1.1.1.8]
Length=352
Score = 75.9 bits (185), Expect = 3e-14, Method: Composition-based stats.
Identities = 37/72 (51%), Positives = 50/72 (69%), Gaps = 1/72 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLA-DTLYDSAGYA 59
CGALKN+VA+A GFC+G+ G NTKAA++RLG+ E+ F F D ++ T +S G A
Sbjct 203 CGALKNIVAVAAGFCDGLSCGDNTKAAVIRLGLMEMIAFANVFCKDSVSIATFLESCGVA 262
Query 60 DVITTVFGGNCR 71
D+ITT +GG R
Sbjct 263 DLITTCYGGRNR 274
> mmu:14555 Gpd1, AI747587, Gdc-1, Gdc1, KIAA4010, mKIAA4010;
glycerol-3-phosphate dehydrogenase 1 (soluble) (EC:1.1.1.8);
K00006 glycerol-3-phosphate dehydrogenase (NAD+) [EC:1.1.1.8]
Length=349
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 37/72 (51%), Positives = 50/72 (69%), Gaps = 1/72 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTF-FGDLLADTLYDSAGYA 59
CGALKN+VA+ GFC+G+G G NTKAA++RLG+ E+ F F G + + T +S G A
Sbjct 200 CGALKNIVAVGAGFCDGLGFGDNTKAAVIRLGLMEMIAFAKLFCSGTVSSATFLESCGVA 259
Query 60 DVITTVFGGNCR 71
D+ITT +GG R
Sbjct 260 DLITTCYGGRNR 271
> dre:550404 gpd1a, zgc:112197; glycerol-3-phosphate dehydrogenase
1a; K00006 glycerol-3-phosphate dehydrogenase (NAD+) [EC:1.1.1.8]
Length=351
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 39/76 (51%), Positives = 52/76 (68%), Gaps = 4/76 (5%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTF--FGDLLADTLYDSAGY 58
CGALKN+VA+ GFC+G+G G NTKAA++RLG+ E+ +F F G + T +S G
Sbjct 201 CGALKNIVAVGAGFCDGLGFGDNTKAAVIRLGLMEMISFARLFCTAGSVSPATFLESCGV 260
Query 59 ADVITTVFGGNCRARR 74
AD+ITT +GG R RR
Sbjct 261 ADLITTCYGG--RNRR 274
> dre:406615 gpd1, wu:fc30a07, zgc:63859; glycerol-3-phosphate
dehydrogenase 1 (soluble) (EC:1.1.1.8); K00006 glycerol-3-phosphate
dehydrogenase (NAD+) [EC:1.1.1.8]
Length=349
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 38/75 (50%), Positives = 51/75 (68%), Gaps = 3/75 (4%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLAD-TLYDSAGYA 59
CGALKN+VA+ GFC+G+G G NTKAA++RLG+ E+ F F ++ T +S G A
Sbjct 200 CGALKNIVAVGAGFCDGLGFGDNTKAAVIRLGLMEMVAFSKLFCKSSVSSATFLESCGVA 259
Query 60 DVITTVFGGNCRARR 74
D+ITT +GG R RR
Sbjct 260 DLITTCYGG--RNRR 272
> xla:444456 MGC83663 protein; K00006 glycerol-3-phosphate dehydrogenase
(NAD+) [EC:1.1.1.8]
Length=349
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 36/72 (50%), Positives = 48/72 (66%), Gaps = 1/72 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFF-GDLLADTLYDSAGYA 59
CGALKN+VA+ GFC+G+ G NTKAA++RLG+ E+ F F G + T +S G A
Sbjct 200 CGALKNIVAVGAGFCDGLDFGDNTKAAVIRLGLMEMVAFAQLFCKGSVCTSTFLESCGVA 259
Query 60 DVITTVFGGNCR 71
D+ITT +GG R
Sbjct 260 DLITTCYGGRNR 271
> sce:YOL059W GPD2, GPD3; Gpd2p (EC:1.1.1.8); K00006 glycerol-3-phosphate
dehydrogenase (NAD+) [EC:1.1.1.8]
Length=440
Score = 73.2 bits (178), Expect = 2e-13, Method: Composition-based stats.
Identities = 41/68 (60%), Positives = 46/68 (67%), Gaps = 1/68 (1%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLY-DSAGYAD 60
GALKNVVALA GF EGMG G N AAI RLG+ EI F FF + +T Y +SAG AD
Sbjct 291 GALKNVVALACGFVEGMGWGNNASAAIQRLGLGEIIKFGRMFFPESKVETYYQESAGVAD 350
Query 61 VITTVFGG 68
+ITT GG
Sbjct 351 LITTCSGG 358
> mmu:333433 Gpd1l, 2210409H23Rik, D9Ertd660e; glycerol-3-phosphate
dehydrogenase 1-like (EC:1.1.1.8); K00006 glycerol-3-phosphate
dehydrogenase (NAD+) [EC:1.1.1.8]
Length=351
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 38/75 (50%), Positives = 50/75 (66%), Gaps = 3/75 (4%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFF-GDLLADTLYDSAGYA 59
CGALKN+VA+ GFC+G+ G NTKAA++RLG+ E+ F F G + T +S G A
Sbjct 202 CGALKNIVAVGAGFCDGLRCGDNTKAAVIRLGLMEMIAFAKIFCKGQVSTATFLESCGVA 261
Query 60 DVITTVFGGNCRARR 74
D+ITT +GG R RR
Sbjct 262 DLITTCYGG--RNRR 274
> hsa:23171 GPD1L, GPD1-L, KIAA0089; glycerol-3-phosphate dehydrogenase
1-like (EC:1.1.1.8); K00006 glycerol-3-phosphate dehydrogenase
(NAD+) [EC:1.1.1.8]
Length=351
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 38/75 (50%), Positives = 50/75 (66%), Gaps = 3/75 (4%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFF-GDLLADTLYDSAGYA 59
CGALKN+VA+ GFC+G+ G NTKAA++RLG+ E+ F F G + T +S G A
Sbjct 202 CGALKNIVAVGAGFCDGLRCGDNTKAAVIRLGLMEMIAFARIFCKGQVSTATFLESCGVA 261
Query 60 DVITTVFGGNCRARR 74
D+ITT +GG R RR
Sbjct 262 DLITTCYGG--RNRR 274
> sce:YDL022W GPD1, DAR1, HOR1, OSG1, OSR5; Gpd1p; K00006 glycerol-3-phosphate
dehydrogenase (NAD+) [EC:1.1.1.8]
Length=391
Score = 72.4 bits (176), Expect = 3e-13, Method: Composition-based stats.
Identities = 39/69 (56%), Positives = 46/69 (66%), Gaps = 1/69 (1%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLY-DSAGYA 59
CGALKNVVAL GF EG+G G N AAI R+G+ EI F FF + +T Y +SAG A
Sbjct 241 CGALKNVVALGCGFVEGLGWGNNASAAIQRVGLGEIIRFGQMFFPESREETYYQESAGVA 300
Query 60 DVITTVFGG 68
D+ITT GG
Sbjct 301 DLITTCAGG 309
> dre:449663 gpd1l, wu:fi13g03, wu:fi45b08, zgc:92580; glycerol-3-phosphate
dehydrogenase 1-like (EC:1.1.1.8); K00006 glycerol-3-phosphate
dehydrogenase (NAD+) [EC:1.1.1.8]
Length=351
Score = 72.4 bits (176), Expect = 4e-13, Method: Composition-based stats.
Identities = 38/76 (50%), Positives = 51/76 (67%), Gaps = 4/76 (5%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGD--LLADTLYDSAGY 58
CGALKN+VA+ GFC+G+ G NTKAA++RLG+ E+ F F D + + T +S G
Sbjct 201 CGALKNIVAVGAGFCDGLQCGDNTKAAVIRLGLMEMIAFAKLFSKDDSVSSATFLESCGV 260
Query 59 ADVITTVFGGNCRARR 74
AD+ITT +GG R RR
Sbjct 261 ADLITTCYGG--RNRR 274
> dre:325181 gpd1b, Gpd1, gpd1h, gpd1l, wu:fc58b05, zgc:66051,
zgc:85742; glycerol-3-phosphate dehydrogenase 1b; K00006 glycerol-3-phosphate
dehydrogenase (NAD+) [EC:1.1.1.8]
Length=350
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 35/73 (47%), Positives = 47/73 (64%), Gaps = 2/73 (2%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTF--FGDLLADTLYDSAGY 58
CGALKN+VA+ GFC+G+ G NTKAA++RLG+ E+ F F + T +S G
Sbjct 200 CGALKNIVAVGAGFCDGLSFGDNTKAAVIRLGLMEMIAFARLFCTASPVSPATFLESCGV 259
Query 59 ADVITTVFGGNCR 71
AD+ITT +GG R
Sbjct 260 ADLITTCYGGRNR 272
> pfa:PFL0780w glycerol-3-phosphate dehydrogenase, putative (EC:1.1.1.8);
K00006 glycerol-3-phosphate dehydrogenase (NAD+)
[EC:1.1.1.8]
Length=367
Score = 70.5 bits (171), Expect = 1e-12, Method: Composition-based stats.
Identities = 31/68 (45%), Positives = 45/68 (66%), Gaps = 0/68 (0%)
Query 1 CGALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYAD 60
CGALKN++ LA GFC+G+ TN+K+AI+R G+ E+ F FF + L +S G+AD
Sbjct 214 CGALKNIITLACGFCDGLNLPTNSKSAIIRNGINEMILFGKVFFQKFNENILLESCGFAD 273
Query 61 VITTVFGG 68
+IT+ G
Sbjct 274 IITSFLAG 281
> cpv:cgd2_210 glycerol-3-phosphate dehydrogenase (EC:1.1.1.8);
K00006 glycerol-3-phosphate dehydrogenase (NAD+) [EC:1.1.1.8]
Length=416
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 33/81 (40%), Positives = 48/81 (59%), Gaps = 4/81 (4%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFF---GDLLADTLYDSAGY 58
G LKN++AL G +G+G GTNT AA++RLGV E+ + FF ++ ++S G
Sbjct 213 GGLKNIIALLVGMIQGLGCGTNTVAAVMRLGVLEMILYGSIFFNIRSSIMTRVFFESCGI 272
Query 59 ADVITTVFGG-NCRARRRLNL 78
AD++TT GG N R + L
Sbjct 273 ADLVTTCLGGRNVRGGKAFTL 293
> ath:AT2G40690 GLY1; GLY1; glycerol-3-phosphate dehydrogenase
(NAD+)
Length=420
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 40/72 (55%), Gaps = 2/72 (2%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYADV 61
GALKNV+A+A G +GM G N+ AA++ G EI+ ++ T G T+ +G D+
Sbjct 276 GALKNVLAIAAGIVDGMNLGNNSMAALVSQGCSEIR-WLATKMG-AKPTTITGLSGTGDI 333
Query 62 ITTVFGGNCRAR 73
+ T F R R
Sbjct 334 MLTCFVNLSRNR 345
> eco:b3608 gpsA, ECK3598, JW3583; glycerol-3-phosphate dehydrogenase
(NAD+) (EC:1.1.1.94); K00057 glycerol-3-phosphate dehydrogenase
(NAD(P)+) [EC:1.1.1.94]
Length=339
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Query 2 GALKNVVALAGGFCEGMGQGTNTKAAILRLGVEEIKTFIMTFFGDLLADTLYDSAGYADV 61
GA+KNV+A+ G +G+G G N + A++ G+ E+ D T AG D+
Sbjct 192 GAVKNVIAIGAGMSDGIGFGANARTALITRGLAEMSRLGAALGAD--PATFMGMAGLGDL 249
Query 62 ITTVFGGNCRARR 74
+ T R RR
Sbjct 250 VLTCTDNQSRNRR 262
> eco:b1633 nth, ECK1629, JW1625; DNA glycosylase and apyrimidinic
(AP) lyase (endonuclease III) (EC:4.2.99.18); K10773 endonuclease
III [EC:4.2.99.18]
Length=211
Score = 28.9 bits (63), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 13/21 (61%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 23 NTKAAILRLGVEEIKTFIMTF 43
NT AA+L LGVE +KT+I T
Sbjct 59 NTPAAMLELGVEGVKTYIKTI 79
Lambda K H
0.326 0.142 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2053886800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40