bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5605_orf1
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
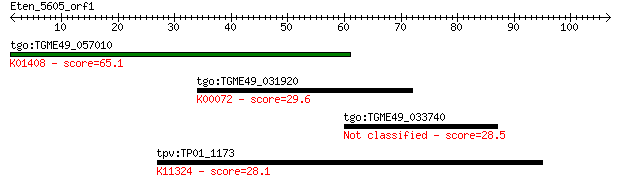
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 65.1 5e-11
tgo:TGME49_031920 sepiapterin reductase, putative (EC:1.1.1.15... 29.6 2.5
tgo:TGME49_033740 hypothetical protein 28.5 5.1
tpv:TP01_1173 hypothetical protein; K11324 DNA methyltransfera... 28.1 7.8
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 44/60 (73%), Gaps = 0/60 (0%)
Query 1 DFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKLSDPSRRMVVKLIADLEPA 60
+F +S +I +H +CF+KRDLE+ YL+ +F+RK L RT+TKL +P+RR+V ++ + P
Sbjct 885 EFGRSWGQIANHGHCFNKRDLELIYLNTEFNRKQLSRTYTKLLEPTRRLVGFVLNHVTPC 944
> tgo:TGME49_031920 sepiapterin reductase, putative (EC:1.1.1.153);
K00072 sepiapterin reductase [EC:1.1.1.153]
Length=303
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 34 HLLRTFTKLSDPSRRMVVKLIADLEPAKE-VTLIGEAKG 71
+ +TF+K++D S + + I++L P+K V LIG +KG
Sbjct 4 YCAKTFSKMADTSVKRSTREISNLFPSKALVLLIGASKG 42
> tgo:TGME49_033740 hypothetical protein
Length=468
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 60 AKEVTLIGEAKGQDAALLRRGDTAEEV 86
AKE+ ++GE +G+ A LL R D E V
Sbjct 320 AKEICIVGEVRGEPAVLLVRLDAPEAV 346
> tpv:TP01_1173 hypothetical protein; K11324 DNA methyltransferase
1-associated protein 1
Length=611
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 17/68 (25%), Positives = 32/68 (47%), Gaps = 0/68 (0%)
Query 27 DNDFSRKHLLRTFTKLSDPSRRMVVKLIADLEPAKEVTLIGEAKGQDAALLRRGDTAEEV 86
D+D R+ +L +++ R M +L++D++ A+ + E K + L+R E
Sbjct 231 DHDRDRRQMLERSYRITPEQREMEAQLLSDIKAAESLLKSEEKKRSEMKRLKRKFNVNES 290
Query 87 VKAPTKLL 94
PT L
Sbjct 291 EVIPTPRL 298
Lambda K H
0.317 0.132 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2012750684
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40