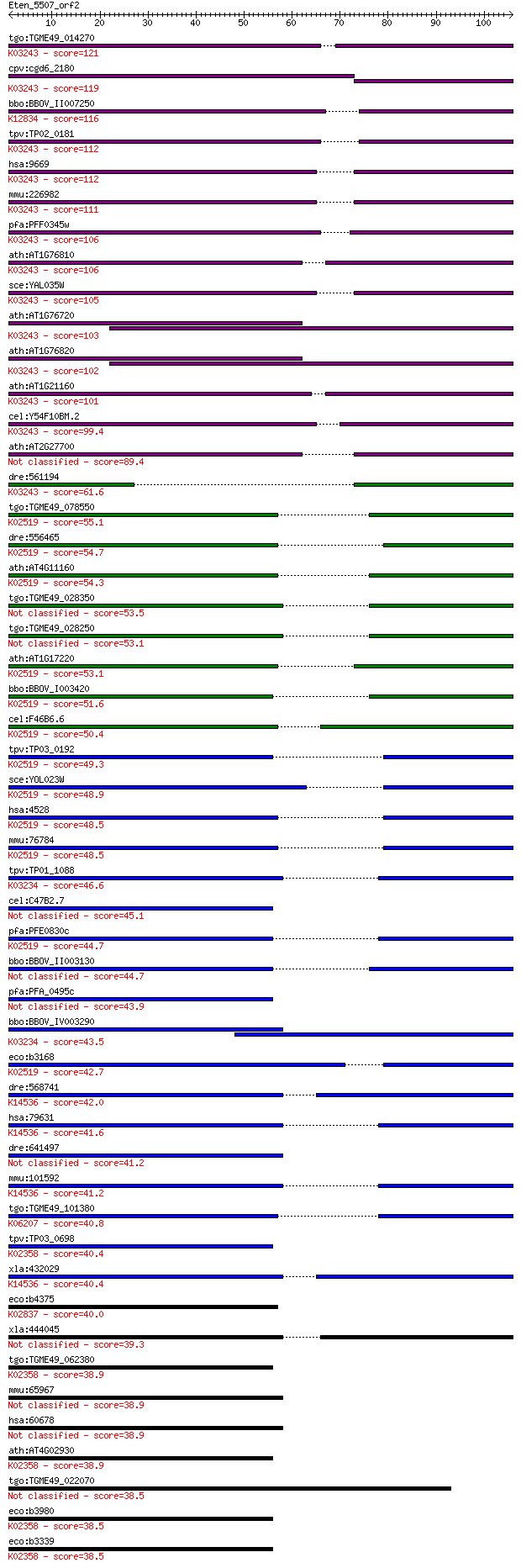
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5507_orf2
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_014270 translation initiation factor IF-2, putative... 121 5e-28
cpv:cgd6_2180 Fun12p GTpase; translation initiation factor IF2... 119 2e-27
bbo:BBOV_II007250 18.m06603; translation initiation factor IF-... 116 2e-26
tpv:TP02_0181 translation initiation factor IF-2; K03243 trans... 112 2e-25
hsa:9669 EIF5B, DKFZp434I036, FLJ10524, IF2, KIAA0741; eukaryo... 112 3e-25
mmu:226982 Eif5b, A030003E17Rik, BC018347, IF2; eukaryotic tra... 111 5e-25
pfa:PFF0345w translation initiation factor IF-2, putative; K03... 106 2e-23
ath:AT1G76810 eukaryotic translation initiation factor 2 famil... 106 2e-23
sce:YAL035W FUN12, yIF2; Fun12p; K03243 translation initiation... 105 5e-23
ath:AT1G76720 GTP binding / GTPase/ translation initiation fac... 103 2e-22
ath:AT1G76820 GTP binding / GTPase; K03243 translation initiat... 102 4e-22
ath:AT1G21160 GTP binding / GTPase/ translation initiation fac... 101 6e-22
cel:Y54F10BM.2 iffb-1; Initiation Factor Five B (eIF5B) family... 99.4 2e-21
ath:AT2G27700 eukaryotic translation initiation factor 2 famil... 89.4 3e-18
dre:561194 im:6912504; si:ch211-254d18.3; K03243 translation i... 61.6 5e-10
tgo:TGME49_078550 elongation factor Tu GTP-binding domain-cont... 55.1 5e-08
dre:556465 mtif2, MGC162191, fb59c03, wu:fb59c03, zgc:162191; ... 54.7 7e-08
ath:AT4G11160 translation initiation factor IF-2, mitochondria... 54.3 9e-08
tgo:TGME49_028350 elongation factor Tu GTP binding domain-cont... 53.5 2e-07
tgo:TGME49_028250 elongation factor Tu GTP-binding domain-cont... 53.1 2e-07
ath:AT1G17220 FUG1; FUG1 (fu-gaeri1); translation initiation f... 53.1 2e-07
bbo:BBOV_I003420 19.m02088; elongation factor Tu GTP binding d... 51.6 7e-07
cel:F46B6.6 hypothetical protein; K02519 translation initiatio... 50.4 1e-06
tpv:TP03_0192 translation initiation factor IF-2; K02519 trans... 49.3 3e-06
sce:YOL023W IFM1; Ifm1p; K02519 translation initiation factor ... 48.9 4e-06
hsa:4528 MTIF2; mitochondrial translational initiation factor ... 48.5 5e-06
mmu:76784 Mtif2, 2310038D14Rik, 2410112O06Rik, IF-2mt; mitocho... 48.5 5e-06
tpv:TP01_1088 elongation factor Tu; K03234 elongation factor 2 46.6
cel:C47B2.7 selb-1; SELB (SelB homolog) translation factor for... 45.1 5e-05
pfa:PFE0830c GTP-binding translation elongation factor tu fami... 44.7 8e-05
bbo:BBOV_II003130 18.m06261; elongation factor Tu GTP binding ... 44.7 8e-05
pfa:PFA_0495c selenocysteine-specific elongation factor SelB h... 43.9 1e-04
bbo:BBOV_IV003290 21.m02927; Elongation factor Tu-like protein... 43.5 2e-04
eco:b3168 infB, ECK3157, gicD, JW3137, ssyG; fused protein cha... 42.7 3e-04
dre:568741 Elongation FacTor family member (eft-2)-like; K1453... 42.0 5e-04
hsa:79631 EFTUD1, FAM42A, FLJ13119, HsT19294, RIA1; elongation... 41.6 7e-04
dre:641497 eefsec, MGC123265, zgc:123265; eukaryotic elongatio... 41.2 8e-04
mmu:101592 Eftud1, 4932434J20Rik, 6030468D11Rik, AI451340, AU0... 41.2 9e-04
tgo:TGME49_101380 GTP-binding protein TypA, putative (EC:2.7.7... 40.8 0.001
tpv:TP03_0698 elongation factor Tu; K02358 elongation factor Tu 40.4 0.001
xla:432029 eftud1, MGC83880; elongation factor Tu GTP binding ... 40.4 0.002
eco:b4375 prfC, ECK4366, JW5873, sra, srb, tos; peptide chain ... 40.0 0.002
xla:444045 eefsec, MGC82641; eukaryotic elongation factor, sel... 39.3 0.003
tgo:TGME49_062380 elongation factor Tu, putative (EC:2.7.7.4);... 38.9 0.004
mmu:65967 Eefsec, Selb, sec; eukaryotic elongation factor, sel... 38.9 0.004
hsa:60678 EEFSEC, EFSEC, SELB; eukaryotic elongation factor, s... 38.9 0.004
ath:AT4G02930 elongation factor Tu, putative / EF-Tu, putative... 38.9 0.004
tgo:TGME49_022070 elongation factor 1-alpha, putative (EC:2.7.... 38.5 0.005
eco:b3980 tufB, ECK3971, JW3943, kirT, pulT; protein chain elo... 38.5 0.005
eco:b3339 tufA, ECK3326, JW3301, kirT, pulT; protein chain elo... 38.5 0.005
> tgo:TGME49_014270 translation initiation factor IF-2, putative
(EC:2.7.7.4); K03243 translation initiation factor 5B
Length=1144
Score = 121 bits (304), Expect = 5e-28, Method: Composition-based stats.
Identities = 52/65 (80%), Positives = 58/65 (89%), Gaps = 0/65 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH+SF NLRARGSSLCDLAIVVVDIMHG+E QTRESL LL+Q+KCP ++ALNKIDRLYGW
Sbjct 620 GHSSFTNLRARGSSLCDLAIVVVDIMHGMEQQTRESLELLKQKKCPFIIALNKIDRLYGW 679
Query 61 KETPW 65
PW
Sbjct 680 NSIPW 684
Score = 68.2 bits (165), Expect = 6e-12, Method: Composition-based stats.
Identities = 31/37 (83%), Positives = 34/37 (91%), Gaps = 0/37 (0%)
Query 69 AQQLLPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
A+ LPGLL+IDTPGH+SF NLRARGSSLCDLAIVVV
Sbjct 606 AELRLPGLLIIDTPGHSSFTNLRARGSSLCDLAIVVV 642
> cpv:cgd6_2180 Fun12p GTpase; translation initiation factor IF2
; K03243 translation initiation factor 5B
Length=896
Score = 119 bits (298), Expect = 2e-27, Method: Composition-based stats.
Identities = 51/72 (70%), Positives = 59/72 (81%), Gaps = 0/72 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF NLR+RGSSLCD+A++VVDIMHGLEPQTRES+ LLR RKCP ++ALNKIDRLYGW
Sbjct 375 GHESFNNLRSRGSSLCDIAVLVVDIMHGLEPQTRESIGLLRSRKCPFIIALNKIDRLYGW 434
Query 61 KETPWGAGAQQL 72
E W + L
Sbjct 435 IEQNWSSSRSTL 446
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 25/33 (75%), Positives = 30/33 (90%), Gaps = 0/33 (0%)
Query 73 LPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+PGLL IDTPGH SF NLR+RGSSLCD+A++VV
Sbjct 365 IPGLLFIDTPGHESFNNLRSRGSSLCDIAVLVV 397
> bbo:BBOV_II007250 18.m06603; translation initiation factor IF-2;
K12834 PHD finger-like domain-containing protein 5A
Length=1033
Score = 116 bits (290), Expect = 2e-26, Method: Composition-based stats.
Identities = 52/66 (78%), Positives = 56/66 (84%), Gaps = 0/66 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF NLRARGSSLCD+AI+VVDIMHGLEPQT ES+ LLR RKC V+ALNKIDRLY W
Sbjct 519 GHESFNNLRARGSSLCDIAILVVDIMHGLEPQTIESIGLLRGRKCYFVIALNKIDRLYKW 578
Query 61 KETPWG 66
K TPW
Sbjct 579 KTTPWA 584
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 27/32 (84%), Positives = 30/32 (93%), Gaps = 0/32 (0%)
Query 74 PGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
PGLL+IDTPGH SF NLRARGSSLCD+AI+VV
Sbjct 510 PGLLIIDTPGHESFNNLRARGSSLCDIAILVV 541
> tpv:TP02_0181 translation initiation factor IF-2; K03243 translation
initiation factor 5B
Length=921
Score = 112 bits (281), Expect = 2e-25, Method: Composition-based stats.
Identities = 48/65 (73%), Positives = 56/65 (86%), Gaps = 0/65 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF NLRARGSSLCD+AI+VVDIMHGLEPQT ES++LL+ RKC ++ALNKIDR+Y W
Sbjct 407 GHESFNNLRARGSSLCDIAILVVDIMHGLEPQTIESINLLKARKCYFIIALNKIDRIYNW 466
Query 61 KETPW 65
TPW
Sbjct 467 SSTPW 471
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 27/32 (84%), Positives = 30/32 (93%), Gaps = 0/32 (0%)
Query 74 PGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
PGLL+IDTPGH SF NLRARGSSLCD+AI+VV
Sbjct 398 PGLLIIDTPGHESFNNLRARGSSLCDIAILVV 429
> hsa:9669 EIF5B, DKFZp434I036, FLJ10524, IF2, KIAA0741; eukaryotic
translation initiation factor 5B; K03243 translation initiation
factor 5B
Length=1220
Score = 112 bits (280), Expect = 3e-25, Method: Composition-based stats.
Identities = 48/64 (75%), Positives = 58/64 (90%), Gaps = 0/64 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF+NLR RGSSLCD+AI+VVDIMHGLEPQT ES++LL+ +KCP +VALNKIDRLY W
Sbjct 705 GHESFSNLRNRGSSLCDIAILVVDIMHGLEPQTIESINLLKSKKCPFIVALNKIDRLYDW 764
Query 61 KETP 64
K++P
Sbjct 765 KKSP 768
Score = 61.6 bits (148), Expect = 5e-10, Method: Composition-based stats.
Identities = 25/33 (75%), Positives = 31/33 (93%), Gaps = 0/33 (0%)
Query 73 LPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+PG+L+IDTPGH SF+NLR RGSSLCD+AI+VV
Sbjct 695 IPGMLIIDTPGHESFSNLRNRGSSLCDIAILVV 727
> mmu:226982 Eif5b, A030003E17Rik, BC018347, IF2; eukaryotic translation
initiation factor 5B; K03243 translation initiation
factor 5B
Length=1216
Score = 111 bits (278), Expect = 5e-25, Method: Composition-based stats.
Identities = 47/64 (73%), Positives = 58/64 (90%), Gaps = 0/64 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF+NLR RGSSLCD+AI+VVDIMHGLEPQT ES+++L+ +KCP +VALNKIDRLY W
Sbjct 701 GHESFSNLRNRGSSLCDIAILVVDIMHGLEPQTIESINILKSKKCPFIVALNKIDRLYDW 760
Query 61 KETP 64
K++P
Sbjct 761 KKSP 764
Score = 61.6 bits (148), Expect = 5e-10, Method: Composition-based stats.
Identities = 25/33 (75%), Positives = 31/33 (93%), Gaps = 0/33 (0%)
Query 73 LPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+PG+L+IDTPGH SF+NLR RGSSLCD+AI+VV
Sbjct 691 IPGMLIIDTPGHESFSNLRNRGSSLCDIAILVV 723
> pfa:PFF0345w translation initiation factor IF-2, putative; K03243
translation initiation factor 5B
Length=977
Score = 106 bits (265), Expect = 2e-23, Method: Composition-based stats.
Identities = 44/65 (67%), Positives = 54/65 (83%), Gaps = 0/65 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF NLR RGSSLCD+AI+V+D+MHGLE QT+ES+ +L+QR CP V+ALNKIDRLY W
Sbjct 465 GHESFYNLRKRGSSLCDIAILVIDLMHGLEQQTKESIQILKQRNCPFVIALNKIDRLYMW 524
Query 61 KETPW 65
+ W
Sbjct 525 NKNDW 529
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/34 (67%), Positives = 29/34 (85%), Gaps = 0/34 (0%)
Query 72 LLPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
L G+++IDTPGH SF NLR RGSSLCD+AI+V+
Sbjct 454 LSKGIMIIDTPGHESFYNLRKRGSSLCDIAILVI 487
> ath:AT1G76810 eukaryotic translation initiation factor 2 family
protein / eIF-2 family protein; K03243 translation initiation
factor 5B
Length=1294
Score = 106 bits (265), Expect = 2e-23, Method: Composition-based stats.
Identities = 48/61 (78%), Positives = 53/61 (86%), Gaps = 0/61 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF NLR+RGSSLCDLAI+VVDIMHGLEPQT ESL+LLR R +VALNK+DRLYGW
Sbjct 777 GHESFTNLRSRGSSLCDLAILVVDIMHGLEPQTIESLNLLRMRNTEFIVALNKVDRLYGW 836
Query 61 K 61
K
Sbjct 837 K 837
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 30/39 (76%), Positives = 34/39 (87%), Gaps = 0/39 (0%)
Query 67 AGAQQLLPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
A A+ +PGLLVIDTPGH SF NLR+RGSSLCDLAI+VV
Sbjct 761 ADAKLKVPGLLVIDTPGHESFTNLRSRGSSLCDLAILVV 799
> sce:YAL035W FUN12, yIF2; Fun12p; K03243 translation initiation
factor 5B
Length=1002
Score = 105 bits (261), Expect = 5e-23, Method: Composition-based stats.
Identities = 47/64 (73%), Positives = 54/64 (84%), Gaps = 0/64 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF+NLR+RGSSLC++AI+V+DIMHGLE QT ES+ LLR RK P VVALNKIDRLY W
Sbjct 479 GHESFSNLRSRGSSLCNIAILVIDIMHGLEQQTIESIKLLRDRKAPFVVALNKIDRLYDW 538
Query 61 KETP 64
K P
Sbjct 539 KAIP 542
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 25/33 (75%), Positives = 32/33 (96%), Gaps = 0/33 (0%)
Query 73 LPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+PGLLVIDTPGH SF+NLR+RGSSLC++AI+V+
Sbjct 469 VPGLLVIDTPGHESFSNLRSRGSSLCNIAILVI 501
> ath:AT1G76720 GTP binding / GTPase/ translation initiation factor;
K03243 translation initiation factor 5B
Length=1191
Score = 103 bits (256), Expect = 2e-22, Method: Composition-based stats.
Identities = 46/61 (75%), Positives = 52/61 (85%), Gaps = 0/61 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF NLR+RGSSLCDLAI+VVDI HGLEPQT ESL+LLR R ++ALNK+DRLYGW
Sbjct 700 GHESFTNLRSRGSSLCDLAILVVDIKHGLEPQTIESLNLLRMRNTEFIIALNKVDRLYGW 759
Query 61 K 61
K
Sbjct 760 K 760
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 39/84 (46%), Positives = 50/84 (59%), Gaps = 5/84 (5%)
Query 22 VVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGWKETPWGAGAQQLLPGLLVIDT 81
++D + G Q E+ + +Q A N +R K A A+ +PGLLVIDT
Sbjct 644 LLDCIRGTNVQEGEAGGITQQIGATYFPAKNIRERTRELK-----ADAKLKVPGLLVIDT 698
Query 82 PGHASFANLRARGSSLCDLAIVVV 105
PGH SF NLR+RGSSLCDLAI+VV
Sbjct 699 PGHESFTNLRSRGSSLCDLAILVV 722
> ath:AT1G76820 GTP binding / GTPase; K03243 translation initiation
factor 5B
Length=1166
Score = 102 bits (253), Expect = 4e-22, Method: Composition-based stats.
Identities = 45/61 (73%), Positives = 52/61 (85%), Gaps = 0/61 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF NLR+RGSSLCDLAI+VVDI HGL+PQT ESL+LLR R ++ALNK+DRLYGW
Sbjct 649 GHESFTNLRSRGSSLCDLAILVVDITHGLQPQTIESLNLLRMRNTEFIIALNKVDRLYGW 708
Query 61 K 61
K
Sbjct 709 K 709
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 39/84 (46%), Positives = 50/84 (59%), Gaps = 5/84 (5%)
Query 22 VVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGWKETPWGAGAQQLLPGLLVIDT 81
++D + G Q E+ + +Q A N +R K A A+ +PGLLVIDT
Sbjct 593 LLDCIRGTNVQEGEAGGITQQIGATYFPAKNIRERTRELK-----ADAKLKVPGLLVIDT 647
Query 82 PGHASFANLRARGSSLCDLAIVVV 105
PGH SF NLR+RGSSLCDLAI+VV
Sbjct 648 PGHESFTNLRSRGSSLCDLAILVV 671
> ath:AT1G21160 GTP binding / GTPase/ translation initiation factor;
K03243 translation initiation factor 5B
Length=1092
Score = 101 bits (251), Expect = 6e-22, Method: Composition-based stats.
Identities = 44/63 (69%), Positives = 55/63 (87%), Gaps = 0/63 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF NLR+RGS+LCDLAI+VVDIM GLEPQT ESL+LLR+R ++ALNK+DRLYGW
Sbjct 569 GHESFTNLRSRGSNLCDLAILVVDIMRGLEPQTIESLNLLRRRNVKFIIALNKVDRLYGW 628
Query 61 KET 63
+++
Sbjct 629 EKS 631
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 28/39 (71%), Positives = 34/39 (87%), Gaps = 0/39 (0%)
Query 67 AGAQQLLPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
A A+ +PG+LVIDTPGH SF NLR+RGS+LCDLAI+VV
Sbjct 553 ANAKLKVPGILVIDTPGHESFTNLRSRGSNLCDLAILVV 591
> cel:Y54F10BM.2 iffb-1; Initiation Factor Five B (eIF5B) family
member (iffb-1); K03243 translation initiation factor 5B
Length=1074
Score = 99.4 bits (246), Expect = 2e-21, Method: Composition-based stats.
Identities = 46/64 (71%), Positives = 52/64 (81%), Gaps = 0/64 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GH SF+NLR RGSSLCD AI+VVDIMHGLEPQT ESL LL + K P V+ALNKIDRLY +
Sbjct 558 GHESFSNLRTRGSSLCDFAILVVDIMHGLEPQTIESLKLLIKGKTPFVIALNKIDRLYEY 617
Query 61 KETP 64
+ P
Sbjct 618 ESNP 621
Score = 62.4 bits (150), Expect = 4e-10, Method: Composition-based stats.
Identities = 26/36 (72%), Positives = 30/36 (83%), Gaps = 0/36 (0%)
Query 70 QQLLPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
Q +PG L+IDTPGH SF+NLR RGSSLCD AI+VV
Sbjct 545 QMKIPGFLIIDTPGHESFSNLRTRGSSLCDFAILVV 580
> ath:AT2G27700 eukaryotic translation initiation factor 2 family
protein / eIF-2 family protein
Length=480
Score = 89.4 bits (220), Expect = 3e-18, Method: Composition-based stats.
Identities = 38/61 (62%), Positives = 48/61 (78%), Gaps = 0/61 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
G+ + NLR+RG LCD AI+VVDIMHGLEPQT E L+LLR + ++ALNK+DRLYGW
Sbjct 107 GYEFYTNLRSRGLGLCDFAILVVDIMHGLEPQTIECLNLLRMKNTEFIIALNKVDRLYGW 166
Query 61 K 61
+
Sbjct 167 R 167
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 19/33 (57%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 73 LPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+P L IDTPG+ + NLR+RG LCD AI+VV
Sbjct 97 VPRPLFIDTPGYEFYTNLRSRGLGLCDFAILVV 129
> dre:561194 im:6912504; si:ch211-254d18.3; K03243 translation
initiation factor 5B
Length=1166
Score = 61.6 bits (148), Expect = 5e-10, Method: Composition-based stats.
Identities = 25/33 (75%), Positives = 31/33 (93%), Gaps = 0/33 (0%)
Query 73 LPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+PG+L+IDTPGH SF+NLR RGSSLCD+AI+VV
Sbjct 704 IPGMLIIDTPGHESFSNLRNRGSSLCDIAILVV 736
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 21/26 (80%), Positives = 24/26 (92%), Gaps = 0/26 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIM 26
GH SF+NLR RGSSLCD+AI+VVDIM
Sbjct 714 GHESFSNLRNRGSSLCDIAILVVDIM 739
> tgo:TGME49_078550 elongation factor Tu GTP-binding domain-containing
protein (EC:2.7.7.4 2.1.1.86); K02519 translation initiation
factor IF-2
Length=2528
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 40/56 (71%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GHA+FA++R RG++ DL ++VV G++ QT E L ++R+ P +VALNK+D+
Sbjct 1974 GHAAFASMRKRGAAATDLCVLVVAADEGVKAQTLECLDIIRKSGTPWIVALNKVDK 2029
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 24/30 (80%), Gaps = 0/30 (0%)
Query 76 LLVIDTPGHASFANLRARGSSLCDLAIVVV 105
L IDTPGHA+FA++R RG++ DL ++VV
Sbjct 1967 LTFIDTPGHAAFASMRKRGAAATDLCVLVV 1996
> dre:556465 mtif2, MGC162191, fb59c03, wu:fb59c03, zgc:162191;
mitochondrial translational initiation factor 2; K02519 translation
initiation factor IF-2
Length=705
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GHA+F+ +RARG+ + D+ I+VV G+ QT ES+ L ++ K P +VA+NK D+
Sbjct 215 GHAAFSAMRARGTMITDIVILVVAADDGVMKQTIESIELAKKAKVPIIVAVNKCDK 270
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 15/27 (55%), Positives = 23/27 (85%), Gaps = 0/27 (0%)
Query 79 IDTPGHASFANLRARGSSLCDLAIVVV 105
+DTPGHA+F+ +RARG+ + D+ I+VV
Sbjct 211 LDTPGHAAFSAMRARGTMITDIVILVV 237
> ath:AT4G11160 translation initiation factor IF-2, mitochondrial,
putative; K02519 translation initiation factor IF-2
Length=743
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 24/56 (42%), Positives = 39/56 (69%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GHA+F+ +RARG+++ D+ ++VV G+ PQT E+++ R P VVA+NK D+
Sbjct 277 GHAAFSEMRARGAAVTDIVVLVVAADDGVMPQTLEAIAHARSANVPVVVAINKCDK 332
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 25/30 (83%), Gaps = 0/30 (0%)
Query 76 LLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+ +DTPGHA+F+ +RARG+++ D+ ++VV
Sbjct 270 ITFLDTPGHAAFSEMRARGAAVTDIVVLVV 299
> tgo:TGME49_028350 elongation factor Tu GTP binding domain-containing
protein (EC:2.7.7.4)
Length=2427
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/57 (45%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRL 57
GHA+F +R R +S DLA++VV + G++PQT S+ + R P VVAL K RL
Sbjct 1536 GHAAFEQMRERATSGADLAVLVVAVDEGVQPQTARSIQMCRAANVPVVVALTKRTRL 1592
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 76 LLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+ +DTPGHA+F +R R +S DLA++VV
Sbjct 1529 VTFLDTPGHAAFEQMRERATSGADLAVLVV 1558
> tgo:TGME49_028250 elongation factor Tu GTP-binding domain-containing
protein (EC:3.1.4.12)
Length=1111
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/57 (43%), Positives = 36/57 (63%), Gaps = 0/57 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRL 57
GHA+FA +R R S+ DLA +VVD+ G QT E L L + P ++ +NK+DR+
Sbjct 273 GHATFAAMRGRASACADLACIVVDVTEGQRLQTDEVLRLADEFDLPLLIVINKVDRI 329
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 18/30 (60%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 76 LLVIDTPGHASFANLRARGSSLCDLAIVVV 105
L IDTPGHA+FA +R R S+ DLA +VV
Sbjct 266 LTFIDTPGHATFAAMRGRASACADLACIVV 295
> ath:AT1G17220 FUG1; FUG1 (fu-gaeri1); translation initiation
factor; K02519 translation initiation factor IF-2
Length=1026
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 24/56 (42%), Positives = 38/56 (67%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GH +F +RARG+ + D+AI+VV G+ PQT E+++ + P V+A+NKID+
Sbjct 561 GHEAFGAMRARGARVTDIAIIVVAADDGIRPQTNEAIAHAKAAAVPIVIAINKIDK 616
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 73 LPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
L + +DTPGH +F +RARG+ + D+AI+VV
Sbjct 551 LQSCVFLDTPGHEAFGAMRARGARVTDIAIIVV 583
> bbo:BBOV_I003420 19.m02088; elongation factor Tu GTP binding
domain containing protein; K02519 translation initiation factor
IF-2
Length=893
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 24/55 (43%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKID 55
GHA+F +R RG D+ I+VV G+ PQT E++ L+++ P +VA+NKID
Sbjct 421 GHAAFGRMRDRGVRCADIVILVVAADDGVMPQTMEAIDLIKRDCLPCIVAVNKID 475
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 76 LLVIDTPGHASFANLRARGSSLCDLAIVVV 105
L+ +DTPGHA+F +R RG D+ I+VV
Sbjct 414 LVFMDTPGHAAFGRMRDRGVRCADIVILVV 443
> cel:F46B6.6 hypothetical protein; K02519 translation initiation
factor IF-2
Length=702
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GHA+FA++RARG+ D+ ++VV G++ QT +S+ + VVA+NKID+
Sbjct 213 GHAAFASMRARGAKGADIVVLVVAADDGVKEQTAQSIKFAKDANVQLVVAVNKIDK 268
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 20/42 (47%), Positives = 30/42 (71%), Gaps = 2/42 (4%)
Query 66 GAGAQQLLPGLLV--IDTPGHASFANLRARGSSLCDLAIVVV 105
GA + +L G V +DTPGHA+FA++RARG+ D+ ++VV
Sbjct 194 GAFSVELTKGRRVTFLDTPGHAAFASMRARGAKGADIVVLVV 235
> tpv:TP03_0192 translation initiation factor IF-2; K02519 translation
initiation factor IF-2
Length=956
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 23/55 (41%), Positives = 34/55 (61%), Gaps = 0/55 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKID 55
GHA+F +R RG S DL I+VV G+ PQT E + L+++ + A+NK+D
Sbjct 461 GHAAFLTMRERGVSCTDLVILVVAADDGVMPQTLECIDLIKRFNLRVIAAINKVD 515
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 17/27 (62%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 79 IDTPGHASFANLRARGSSLCDLAIVVV 105
IDTPGHA+F +R RG S DL I+VV
Sbjct 457 IDTPGHAAFLTMRERGVSCTDLVILVV 483
> sce:YOL023W IFM1; Ifm1p; K02519 translation initiation factor
IF-2
Length=676
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRLYGW 60
GHA+F +R RG+++ D+ ++VV + L PQT E++ + ++A+ KIDR+
Sbjct 203 GHAAFLKMRERGANITDIIVLVVSVEDSLMPQTLEAIKHAKNSGNEMIIAITKIDRIPQP 262
Query 61 KE 62
KE
Sbjct 263 KE 264
Score = 37.7 bits (86), Expect = 0.009, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 79 IDTPGHASFANLRARGSSLCDLAIVVV 105
+DTPGHA+F +R RG+++ D+ ++VV
Sbjct 199 LDTPGHAAFLKMRERGANITDIIVLVV 225
> hsa:4528 MTIF2; mitochondrial translational initiation factor
2; K02519 translation initiation factor IF-2
Length=727
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GHA+F+ +RARG+ + D+ ++VV G+ QT ES+ + + P ++A+NK D+
Sbjct 237 GHAAFSAMRARGAQVTDIVVLVVAADDGVMKQTVESIQHAKDAQVPIILAVNKCDK 292
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 23/27 (85%), Gaps = 0/27 (0%)
Query 79 IDTPGHASFANLRARGSSLCDLAIVVV 105
+DTPGHA+F+ +RARG+ + D+ ++VV
Sbjct 233 LDTPGHAAFSAMRARGAQVTDIVVLVV 259
> mmu:76784 Mtif2, 2310038D14Rik, 2410112O06Rik, IF-2mt; mitochondrial
translational initiation factor 2; K02519 translation
initiation factor IF-2
Length=727
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 37/56 (66%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GHA+F+ +RARG+ + D+ ++VV G+ QT ES+ + + P ++A+NK D+
Sbjct 237 GHAAFSAMRARGAQVTDIVVLVVAADDGVMKQTVESIQHAKDAEVPIILAINKCDK 292
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 23/27 (85%), Gaps = 0/27 (0%)
Query 79 IDTPGHASFANLRARGSSLCDLAIVVV 105
+DTPGHA+F+ +RARG+ + D+ ++VV
Sbjct 233 LDTPGHAAFSAMRARGAQVTDIVVLVV 259
> tpv:TP01_1088 elongation factor Tu; K03234 elongation factor
2
Length=1210
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 23/57 (40%), Positives = 34/57 (59%), Gaps = 0/57 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRL 57
GH F+ + + LCD A++VVD++ G+ PQTR L +V+ LNKID+L
Sbjct 103 GHVDFSIEVSTAARLCDGALLVVDVVEGICPQTRAVLRQAWLENVKTVLILNKIDKL 159
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 78 VIDTPGHASFANLRARGSSLCDLAIVVV 105
+ID+PGH F+ + + LCD A++VV
Sbjct 98 LIDSPGHVDFSIEVSTAARLCDGALLVV 125
> cel:C47B2.7 selb-1; SELB (SelB homolog) translation factor for
selenocysteine incorporation family member (selb-1)
Length=500
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 22/57 (38%), Positives = 36/57 (63%), Gaps = 3/57 (5%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPS--VVALNKID 55
GH+ S++ D+AIV++D++ G++PQT E L LL + CP+ ++ LNK D
Sbjct 81 GHSGLIRAVLAASTVFDMAIVIIDVVAGIQPQTAEHL-LLASKFCPNRVIIVLNKCD 136
> pfa:PFE0830c GTP-binding translation elongation factor tu family
protein, putative; K02519 translation initiation factor
IF-2
Length=1610
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 18/55 (32%), Positives = 35/55 (63%), Gaps = 0/55 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKID 55
GH +F +R+RG + DL+I+V+ G++ QT E + L+++ ++A+ K+D
Sbjct 1049 GHEAFMPMRSRGVKISDLSILVISGDEGIQEQTVECIKLIKEFNIKIIIAITKVD 1103
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 78 VIDTPGHASFANLRARGSSLCDLAIVVV 105
++DTPGH +F +R+RG + DL+I+V+
Sbjct 1044 LVDTPGHEAFMPMRSRGVKISDLSILVI 1071
> bbo:BBOV_II003130 18.m06261; elongation factor Tu GTP binding
domain containing protein
Length=995
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 33/55 (60%), Gaps = 0/55 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKID 55
GH F +R G+++ D+A++V+D + GL+ QT E + L + P ++A K D
Sbjct 542 GHEVFDAMRRCGATIADIALIVIDSIEGLKEQTIECIKLCKSLSIPFIIAATKCD 596
Score = 35.4 bits (80), Expect = 0.046, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 76 LLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+ VIDTPGH F +R G+++ D+A++V+
Sbjct 535 MTVIDTPGHEVFDAMRRCGATIADIALIVI 564
> pfa:PFA_0495c selenocysteine-specific elongation factor SelB
homologue, putative
Length=934
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 0/55 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKID 55
GH S GS + D+ I+V+DI G++ QT E L L + C ++ LNKID
Sbjct 145 GHHSLLKSIIMGSEITDIIILVIDINKGIQKQTIECLVLCKIINCDIIIVLNKID 199
> bbo:BBOV_IV003290 21.m02927; Elongation factor Tu-like protein;
K03234 elongation factor 2
Length=1222
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 35/57 (61%), Gaps = 0/57 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRL 57
GH F+ A + LCD A+++VD++ G+ PQT+ L + +V+ LNK+D+L
Sbjct 103 GHVDFSVEVATAARLCDGALLIVDVVEGICPQTKAVLRQAWRESVRTVLVLNKMDKL 159
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 31/62 (50%), Gaps = 4/62 (6%)
Query 48 VVALNKIDRLYGWKETPWGAGAQQLL---PGLL-VIDTPGHASFANLRARGSSLCDLAIV 103
+ + I LY +T G +L P ++ ++D PGH F+ A + LCD A++
Sbjct 64 TIKSSSISLLYSASDTSNRTGCNRLFNDQPCIINLVDCPGHVDFSVEVATAARLCDGALL 123
Query 104 VV 105
+V
Sbjct 124 IV 125
> eco:b3168 infB, ECK3157, gicD, JW3137, ssyG; fused protein chain
initiation factor 2, IF2: membrane protein/conserved protein;
K02519 translation initiation factor IF-2
Length=890
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 29/83 (34%), Positives = 44/83 (53%), Gaps = 13/83 (15%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR---- 56
GHA+F ++RARG+ D+ ++VV G+ PQT E++ + + P VVA+NKID+
Sbjct 447 GHAAFTSMRARGAQATDIVVLVVAADDGVMPQTIEAIQHAKAAQVPVVVAVNKIDKPEAD 506
Query 57 ---------LYGWKETPWGAGAQ 70
YG WG +Q
Sbjct 507 PDRVKNELSQYGILPEEWGGESQ 529
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 14/27 (51%), Positives = 22/27 (81%), Gaps = 0/27 (0%)
Query 79 IDTPGHASFANLRARGSSLCDLAIVVV 105
+DTPGHA+F ++RARG+ D+ ++VV
Sbjct 443 LDTPGHAAFTSMRARGAQATDIVVLVV 469
> dre:568741 Elongation FacTor family member (eft-2)-like; K14536
ribosome assembly protein 1 [EC:3.6.5.-]
Length=1115
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/57 (42%), Positives = 32/57 (56%), Gaps = 0/57 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRL 57
GH F++ + LCD AIVVVD + G+ PQT+ L V+ +NKIDRL
Sbjct 95 GHVDFSSEVSTAVRLCDGAIVVVDAVEGVCPQTQVVLRQAWLENIRPVLVINKIDRL 151
Score = 31.6 bits (70), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query 65 WGAGAQQLLPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+ G + L L ID+PGH F++ + LCD AIVVV
Sbjct 79 FATGGVEFLINL--IDSPGHVDFSSEVSTAVRLCDGAIVVV 117
> hsa:79631 EFTUD1, FAM42A, FLJ13119, HsT19294, RIA1; elongation
factor Tu GTP binding domain containing 1; K14536 ribosome
assembly protein 1 [EC:3.6.5.-]
Length=1069
Score = 41.6 bits (96), Expect = 7e-04, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRL 57
GH F++ + +CD I+VVD + G+ PQT+ L V+ +NKIDRL
Sbjct 44 GHVDFSSEVSTAVRICDGCIIVVDAVEGVCPQTQAVLRQAWLENIRPVLVINKIDRL 100
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 78 VIDTPGHASFANLRARGSSLCDLAIVVV 105
+ID+PGH F++ + +CD I+VV
Sbjct 39 LIDSPGHVDFSSEVSTAVRICDGCIIVV 66
> dre:641497 eefsec, MGC123265, zgc:123265; eukaryotic elongation
factor, selenocysteine-tRNA-specific
Length=576
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 25/58 (43%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPS-VVALNKIDRL 57
GHAS G+ + DL ++VVD++ G++ QT E L L+ Q C VV LNK D L
Sbjct 87 GHASLIRTIIGGAQIIDLMMLVVDVVKGMQTQTAECL-LIGQLTCSRMVVILNKTDLL 143
> mmu:101592 Eftud1, 4932434J20Rik, 6030468D11Rik, AI451340, AU019507,
AU022896, D7Ertd791e; elongation factor Tu GTP binding
domain containing 1; K14536 ribosome assembly protein 1
[EC:3.6.5.-]
Length=1127
Score = 41.2 bits (95), Expect = 9e-04, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDRL 57
GH F++ + +CD I+VVD + G+ PQT+ L V+ +NKIDRL
Sbjct 95 GHVDFSSEVSTAVRICDGCIIVVDAVEGVCPQTQAVLRQAWLENIRPVLVINKIDRL 151
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 78 VIDTPGHASFANLRARGSSLCDLAIVVV 105
+ID+PGH F++ + +CD I+VV
Sbjct 90 LIDSPGHVDFSSEVSTAVRICDGCIIVV 117
> tgo:TGME49_101380 GTP-binding protein TypA, putative (EC:2.7.7.4);
K06207 GTP-binding protein
Length=749
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/56 (41%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GHA F+ R L D ++VVD + G +PQTR L Q +VV +NK+DR
Sbjct 109 GHADFSGEVERVMHLADGVLLVVDAVEGCKPQTRVVLQKALQAGLRAVVVVNKVDR 164
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 78 VIDTPGHASFANLRARGSSLCDLAIVVV 105
+IDTPGHA F+ R L D ++VV
Sbjct 104 IIDTPGHADFSGEVERVMHLADGVLLVV 131
> tpv:TP03_0698 elongation factor Tu; K02358 elongation factor
Tu
Length=445
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 25/56 (44%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPS-VVALNKID 55
GHA + G++ D AI+VV G PQTRE + L RQ P VV LNK+D
Sbjct 134 GHADYIKNMISGAAQMDGAILVVSAPDGPMPQTREHILLARQIGVPRLVVYLNKMD 189
> xla:432029 eftud1, MGC83880; elongation factor Tu GTP binding
domain containing 1; K14536 ribosome assembly protein 1 [EC:3.6.5.-]
Length=310
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 35/60 (58%), Gaps = 6/60 (10%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQ---RKCPSVVALNKIDRL 57
GH F++ + LCD I+VVD + G+ PQT+ ++LRQ V+ +NKIDRL
Sbjct 95 GHVDFSSEVSTAVRLCDGCIIVVDSVEGVCPQTQ---AVLRQAWLENIRPVLVINKIDRL 151
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query 65 WGAGAQQLLPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
+ G ++ L L ID+PGH F++ + LCD I+VV
Sbjct 79 YKDGEEEYLINL--IDSPGHVDFSSEVSTAVRLCDGCIIVV 117
> eco:b4375 prfC, ECK4366, JW5873, sra, srb, tos; peptide chain
release factor RF-3; K02837 peptide chain release factor 3
Length=529
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 31/56 (55%), Gaps = 0/56 (0%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVALNKIDR 56
GH F+ R + D ++V+D G+E +TR+ + + R R P + +NK+DR
Sbjct 91 GHEDFSEDTYRTLTAVDCCLMVIDAAKGVEDRTRKLMEVTRLRDTPILTFMNKLDR 146
> xla:444045 eefsec, MGC82641; eukaryotic elongation factor, selenocysteine-tRNA-specific
Length=575
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 23/58 (39%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPS-VVALNKIDRL 57
GHAS G+ + DL ++V+D+ G++ Q+ E L ++ Q C VV LNKID L
Sbjct 83 GHASLIRTIIGGAQIIDLMMLVIDVTKGMQTQSAECL-VIGQIACNKMVVVLNKIDLL 139
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query 66 GAGAQQLLPGLLVIDTPGHASFANLRARGSSLCDLAIVVV 105
G G Q+L ++D PGHAS G+ + DL ++V+
Sbjct 68 GTGYQRL--QFTLVDCPGHASLIRTIIGGAQIIDLMMLVI 105
> tgo:TGME49_062380 elongation factor Tu, putative (EC:2.7.7.4);
K02358 elongation factor Tu
Length=552
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 24/56 (42%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPS-VVALNKID 55
GHA + G++ D AI+VV G PQTRE + L +Q P VV LNK+D
Sbjct 237 GHADYVKNMITGAAQMDGAILVVSAYDGPMPQTREHILLSKQVGVPRLVVYLNKMD 292
> mmu:65967 Eefsec, Selb, sec; eukaryotic elongation factor, selenocysteine-tRNA-specific
Length=583
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 23/58 (39%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPS-VVALNKIDRL 57
GHAS G+ + DL ++V+D+ G++ Q+ E L ++ Q C VV LNKID L
Sbjct 81 GHASLIRTIIGGAQIIDLMMLVIDVTKGMQTQSAECL-VIGQIACQKLVVVLNKIDLL 137
> hsa:60678 EEFSEC, EFSEC, SELB; eukaryotic elongation factor,
selenocysteine-tRNA-specific
Length=596
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 23/58 (39%), Positives = 34/58 (58%), Gaps = 2/58 (3%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPS-VVALNKIDRL 57
GHAS G+ + DL ++V+D+ G++ Q+ E L ++ Q C VV LNKID L
Sbjct 95 GHASLIRTIIGGAQIIDLMMLVIDVTKGMQTQSAECL-VIGQIACQKLVVVLNKIDLL 151
> ath:AT4G02930 elongation factor Tu, putative / EF-Tu, putative;
K02358 elongation factor Tu
Length=454
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVA-LNKID 55
GHA + G++ D I+VV G PQT+E + L RQ PS+V LNK+D
Sbjct 139 GHADYVKNMITGAAQMDGGILVVSGPDGPMPQTKEHILLARQVGVPSLVCFLNKVD 194
> tgo:TGME49_022070 elongation factor 1-alpha, putative (EC:2.7.7.4)
Length=645
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 34/102 (33%), Positives = 49/102 (48%), Gaps = 14/102 (13%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMH---GLEPQTRESLSLLRQRKCPSV----VALNK 53
GH F G+ L D+A++VVD G + QT+E L + R C + VALNK
Sbjct 251 GHREFVCNMLGGAVLADVALLVVDTARFEAGFDGQTKEHLLIAR---CLGIQHFLVALNK 307
Query 54 IDRLYGWKETPWGAGAQQL---LPGLLVIDTPGHASFANLRA 92
+D L W E + ++L + GL + PG SF + A
Sbjct 308 MDEL-AWSEEMYAKTVERLRAYMVGLEMNCAPGQISFVPISA 348
> eco:b3980 tufB, ECK3971, JW3943, kirT, pulT; protein chain elongation
factor EF-Tu (duplicate of TufA); K02358 elongation
factor Tu
Length=394
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 30/56 (53%), Gaps = 1/56 (1%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVA-LNKID 55
GHA + G++ D AI+VV G PQTRE + L RQ P ++ LNK D
Sbjct 84 GHADYVKNMITGAAQMDGAILVVAATDGPMPQTREHILLGRQVGVPYIIVFLNKCD 139
> eco:b3339 tufA, ECK3326, JW3301, kirT, pulT; protein chain elongation
factor EF-Tu (duplicate of TufB); K02358 elongation
factor Tu
Length=394
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 23/56 (41%), Positives = 30/56 (53%), Gaps = 1/56 (1%)
Query 1 GHASFANLRARGSSLCDLAIVVVDIMHGLEPQTRESLSLLRQRKCPSVVA-LNKID 55
GHA + G++ D AI+VV G PQTRE + L RQ P ++ LNK D
Sbjct 84 GHADYVKNMITGAAQMDGAILVVAATDGPMPQTREHILLGRQVGVPYIIVFLNKCD 139
Lambda K H
0.323 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017521068
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40