bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
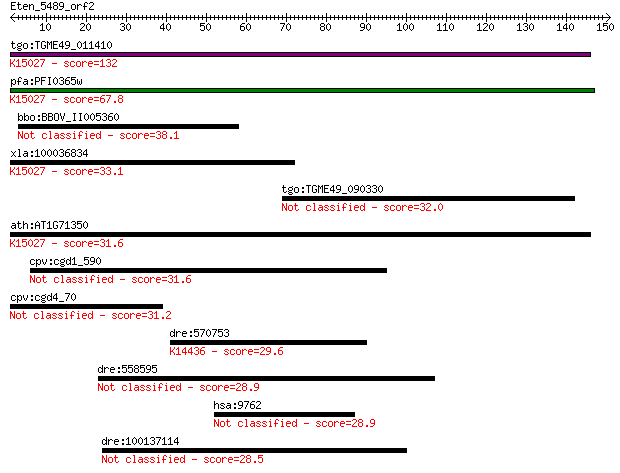
Query= Eten_5489_orf2
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_011410 hypothetical protein ; K15027 translation in... 132 5e-31
pfa:PFI0365w translation initiation factor SUI1, putative; K15... 67.8 1e-11
bbo:BBOV_II005360 18.m10002; hypothetical protein 38.1 0.010
xla:100036834 eif2d, lgtn; eukaryotic translation initiation f... 33.1 0.39
tgo:TGME49_090330 chloride channel protein, putative (EC:2.1.1... 32.0 0.80
ath:AT1G71350 eukaryotic translation initiation factor SUI1 fa... 31.6 0.97
cpv:cgd1_590 hypothetical protein 31.6 1.1
cpv:cgd4_70 hypothetical protein 31.2 1.4
dre:570753 chd6; chromodomain helicase DNA binding protein 6; ... 29.6 3.5
dre:558595 A-kinase anchor protein 11-like 28.9 5.7
hsa:9762 ProSAPiP1, KIAA0552; ProSAPiP1 protein 28.9 6.0
dre:100137114 zgc:173638 28.5 7.8
> tgo:TGME49_011410 hypothetical protein ; K15027 translation
initiation factor 2D
Length=748
Score = 132 bits (331), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 70/150 (46%), Positives = 98/150 (65%), Gaps = 6/150 (4%)
Query 1 DIRRSSARKLGKFIQGATKKKLVQTKEQRGIVSVVKINRSHPDYCKHTPVPESQKKKLVE 60
D+R++S +KL KF+Q +K KL+QTKE RG V++VK+NRSHP + ++ P+PE KKK++E
Sbjct 409 DVRKTSHKKLVKFVQFLSKTKLLQTKETRGAVAMVKVNRSHPQFLEYKPLPEKTKKKILE 468
Query 61 RAGASAAATSPTASGAVGAGGEEGTRDAPNAA-----GTARSGDEGPLVFEFCAPPAKCC 115
R A AA S ++ + G G E G R P A G A D GPLV EFC PP K
Sbjct 469 RVAAEEAAASASSPTSAG-GEETGDRRQPGRAVAGESGAANGSDSGPLVLEFCTPPQKLT 527
Query 116 RIFAAVEVRTGRNVFFTAEEYRQVLVKYFE 145
++F AV V+TG++ FFT E ++VL +Y +
Sbjct 528 KVFHAVNVQTGKDTFFTFAEAKEVLSRYLK 557
> pfa:PFI0365w translation initiation factor SUI1, putative; K15027
translation initiation factor 2D
Length=818
Score = 67.8 bits (164), Expect = 1e-11, Method: Composition-based stats.
Identities = 41/147 (27%), Positives = 69/147 (46%), Gaps = 15/147 (10%)
Query 1 DIRRSSARKLGKFIQGATKKKLVQTKEQRGIVSVVKINRSHPDYCKHTPVPESQKKKL-V 59
D+++SS +KL KFIQ +K KL++ KE R IVS+V I R H Y + P+ KKK +
Sbjct 513 DVKKSSYKKLTKFIQHYSKMKLLKIKENRNIVSIVNIERQHALYKSYEPINVELKKKYDI 572
Query 60 ERAGASAAATSPTASGAVGAGGEEGTRDAPNAAGTARSGDEGPLVFEFCAPPAKCCRIFA 119
E + + ++ N T S ++G V EF P K I
Sbjct 573 ENEQTNLSINE--------------NKNKINKTPTNHSTNKGAQVLEFYIPSNKTLNIIQ 618
Query 120 AVEVRTGRNVFFTAEEYRQVLVKYFEV 146
++E + ++ +F + + + Y ++
Sbjct 619 SIENKVDKSSYFNISQLKDIFRSYIKL 645
> bbo:BBOV_II005360 18.m10002; hypothetical protein
Length=465
Score = 38.1 bits (87), Expect = 0.010, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 3 RRSSARKLGKFIQGATKKKLVQTKEQRGIVSVVKINRSHPDYCKHTPVPESQKKK 57
R SS +KL K Q +K+ L+ K+ RG VV INR H Y + P Q K+
Sbjct 245 RNSSFKKLIKMFQTWSKEGLILMKDIRGTGVVVNINRGHTVYVQFKPWQLPQNKE 299
> xla:100036834 eif2d, lgtn; eukaryotic translation initiation
factor 2D; K15027 translation initiation factor 2D
Length=583
Score = 33.1 bits (74), Expect = 0.39, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 42/72 (58%), Gaps = 2/72 (2%)
Query 1 DIRRSSARKLGKFIQGATKKKLVQTKE-QRGIVSVVKINRSHPDYCKHTPVPESQKKKLV 59
DI++SS +KL KF+Q ++K++Q KE +G+ S+V+I+ HPD + P+S LV
Sbjct 305 DIKKSSYKKLSKFLQTMQQRKVLQVKELSKGVESIVEIDWKHPD-VRSFVAPDSVPVDLV 363
Query 60 ERAGASAAATSP 71
+ SP
Sbjct 364 VSESINGEGESP 375
> tgo:TGME49_090330 chloride channel protein, putative (EC:2.1.1.86)
Length=1590
Score = 32.0 bits (71), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 32/73 (43%), Gaps = 8/73 (10%)
Query 69 TSPTASGAVGAGGEEGTRDAPNAAGTARSGDEGPLVFEFCAPPAKCCRIFAAVEVRTGRN 128
+SP A VGA A AAGT R+ +VFE + + AV + G N
Sbjct 887 SSPAAYALVGA--------ASLAAGTTRTISVSVIVFELTGKLSHMIPVLLAVLIAYGVN 938
Query 129 VFFTAEEYRQVLV 141
FFT Y +L+
Sbjct 939 GFFTISIYDVMLL 951
> ath:AT1G71350 eukaryotic translation initiation factor SUI1
family protein; K15027 translation initiation factor 2D
Length=597
Score = 31.6 bits (70), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 61/147 (41%), Gaps = 32/147 (21%)
Query 1 DIRRSSARKLGKFIQGATKKKLVQTKE--QRGIVSVVKINRSHPDYCKHTPVPESQKKKL 58
DI++SS +KL K++Q ++ KE + + ++ +NRSHPDY P +KKK
Sbjct 320 DIKKSSHKKLSKWLQSKASGGMISVKEDKHKKEIVLISVNRSHPDYKSFKP----EKKK- 374
Query 59 VERAGASAAATSPTASGAVGAGGEEGTRDAPNAAGTARSGDEGPLVFEFCAPPAKCCRIF 118
+ + SP GE T A + G + E IF
Sbjct 375 ------AEVSESP---------GERSTAQAQSEKGLE--------IIEVYKSSIHNSAIF 411
Query 119 AAVEVRTGRNVFFTAEEYRQVLVKYFE 145
A+V G +TA E V+ KY E
Sbjct 412 ASVGEDKGN--LYTASEATDVVFKYIE 436
> cpv:cgd1_590 hypothetical protein
Length=2527
Score = 31.6 bits (70), Expect = 1.1, Method: Composition-based stats.
Identities = 28/90 (31%), Positives = 44/90 (48%), Gaps = 3/90 (3%)
Query 6 SARKLGKFIQGATKKKLVQTKEQRGIVSVVKINRSHPDYCKHTPVPESQKK-KLVERAGA 64
++RK GK +GA +KK K+Q+ + K + V ES+ ++ E +
Sbjct 2016 ASRKHGK--KGAGRKKSRSDKKQKAKTNGKKRRLGLGKKLGTSLVKESKPTLEIPETQIS 2073
Query 65 SAAATSPTASGAVGAGGEEGTRDAPNAAGT 94
S+ SPTA+G A G T ++P AGT
Sbjct 2074 SSVPESPTATGTPIATGSPMTPESPTVAGT 2103
> cpv:cgd4_70 hypothetical protein
Length=718
Score = 31.2 bits (69), Expect = 1.4, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 26/39 (66%), Gaps = 1/39 (2%)
Query 1 DIRRSSARKLGKFIQGATKKKLVQTKEQR-GIVSVVKIN 38
DI++SS K+ KF Q +KK ++ K R G++++V IN
Sbjct 402 DIKKSSFVKVQKFFQYYSKKNILLIKHGRGGVLNIVDIN 440
> dre:570753 chd6; chromodomain helicase DNA binding protein 6;
K14436 chromodomain-helicase-DNA-binding protein 6 [EC:3.6.4.12]
Length=2699
Score = 29.6 bits (65), Expect = 3.5, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 24/51 (47%), Gaps = 8/51 (15%)
Query 41 HPDYCKHTPVPESQKKKLVERAG--ASAAATSPTASGAVGAGGEEGTRDAP 89
HP HTP +LV G S+ A+ P +S +G+G EG D P
Sbjct 1444 HPTPAHHTPT------QLVHTHGHHQSSEASEPGSSLGIGSGNREGFLDCP 1488
> dre:558595 A-kinase anchor protein 11-like
Length=1795
Score = 28.9 bits (63), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 37/93 (39%), Gaps = 9/93 (9%)
Query 23 VQTKEQRGIVSVVKINRSHPDYCKHTPVPESQKKKLVERAGASAAATSPTASGAVGAGG- 81
++ +++ G V+ + I R H D K T E +E+A + TS A +G G
Sbjct 1507 IRRQQESGAVTGLDIPRIHIDLEKRTAFAEEMVSSAIEKAKKELSCTSLNADSGIGHDGA 1566
Query 82 --------EEGTRDAPNAAGTARSGDEGPLVFE 106
E T NA T G EG V E
Sbjct 1567 SFAESLTTEIMTSALSNACQTINLGKEGGNVTE 1599
> hsa:9762 ProSAPiP1, KIAA0552; ProSAPiP1 protein
Length=673
Score = 28.9 bits (63), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 52 ESQKKKLVERAGASAAATSPTASGAVGAGGEEGTR 86
ES + K+ +AG +AAA+ + G AGGE GTR
Sbjct 537 ESDEAKMRRQAGVAAAASLVSVDGEAEAGGESGTR 571
> dre:100137114 zgc:173638
Length=456
Score = 28.5 bits (62), Expect = 7.8, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 40/82 (48%), Gaps = 8/82 (9%)
Query 24 QTKEQRGIVSVVKINRSHPD-----YCKHT-PVPESQKKKLVERAGASAAATSPTASGAV 77
Q +E GIV V++ ++ D + +H+ PV E + + G +A+T S +
Sbjct 351 QMEEISGIVKTVQVTQTPEDQTSEEFQQHSHPVLEESSQGSIPCFGFGSASTG--KSSGI 408
Query 78 GAGGEEGTRDAPNAAGTARSGD 99
G+G E + + G+ R+GD
Sbjct 409 GSGFESASTGKSFSFGSTRTGD 430
Lambda K H
0.316 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3134054208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40