bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5459_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
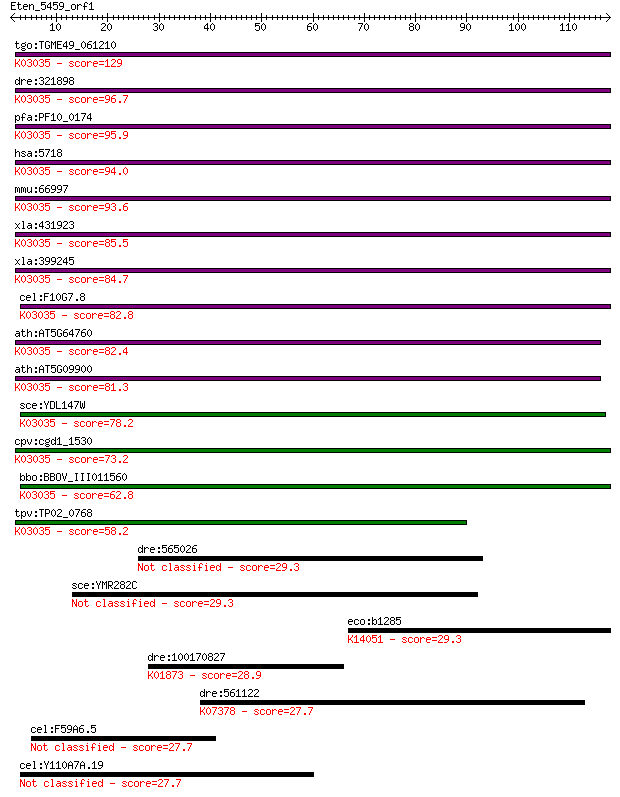
tgo:TGME49_061210 26S proteasome non-ATPase regulatory subunit... 129 3e-30
dre:321898 psmd12, wu:fb38a06, wu:fc34f08, wu:fc49f02; proteas... 96.7 1e-20
pfa:PF10_0174 26S proteasome subunit p55, putative; K03035 26S... 95.9 3e-20
hsa:5718 PSMD12, MGC75406, Rpn5, p55; proteasome (prosome, mac... 94.0 1e-19
mmu:66997 Psmd12, 1500002F15Rik, AI480719, P55; proteasome (pr... 93.6 1e-19
xla:431923 hypothetical protein MGC81129; K03035 26S proteasom... 85.5 4e-17
xla:399245 psmd12, MGC80339; proteasome (prosome, macropain) 2... 84.7 6e-17
cel:F10G7.8 rpn-5; proteasome Regulatory Particle, Non-ATPase-... 82.8 2e-16
ath:AT5G64760 RPN5B; RPN5B (REGULATORY PARTICLE NON-ATPASE SUB... 82.4 3e-16
ath:AT5G09900 EMB2107 (EMBRYO DEFECTIVE 2107); K03035 26S prot... 81.3 6e-16
sce:YDL147W RPN5, NAS5; Essential, non-ATPase regulatory subun... 78.2 5e-15
cpv:cgd1_1530 Rpn5 like 26S proteasomal regulatory subunit 12,... 73.2 2e-13
bbo:BBOV_III011560 17.m07987; PCI domain containing protein; K... 62.8 3e-10
tpv:TP02_0768 26S proteasome subunit p55; K03035 26S proteasom... 58.2 6e-09
dre:565026 tgfbrap1, MGC162302, zgc:162302; transforming growt... 29.3 2.9
sce:YMR282C AEP2, ATP13; Aep2p 29.3 3.2
eco:b1285 gmr, ECK1280, JW1278, yciR; cyclic-di-GMP phosphodie... 29.3 3.6
dre:100170827 vars2, si:ch211-152p11.2; valyl-tRNA synthetase ... 28.9 3.9
dre:561122 nlgn4b, nlgn4a; neuroligin 4b; K07378 neuroligin 27.7 8.9
cel:F59A6.5 hypothetical protein 27.7 9.3
cel:Y110A7A.19 hypothetical protein 27.7 9.9
> tgo:TGME49_061210 26S proteasome non-ATPase regulatory subunit,
putative ; K03035 26S proteasome regulatory subunit N5
Length=485
Score = 129 bits (323), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 64/117 (54%), Positives = 87/117 (74%), Gaps = 1/117 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEA-ADLQVLKVK 60
++VQVETFGAM+R EKT+YILKQM L+LR+ DFIRCQI+SKK+S KLL+ DLQ LK++
Sbjct 189 QEVQVETFGAMERREKTEYILKQMSLVLRRGDFIRCQIISKKISTKLLDNDEDLQDLKIR 248
Query 61 YYELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
YY LM VYYLHE +LD K + I+ T ++ + + L+ + +F++LAPFD E
Sbjct 249 YYSLMIVYYLHEGMILDCCKAYQSIFITPSVQQKEDQWIKSLQCYILFLLLAPFDDE 305
> dre:321898 psmd12, wu:fb38a06, wu:fc34f08, wu:fc49f02; proteasome
(prosome, macropain) 26S subunit, non-ATPase, 12; K03035
26S proteasome regulatory subunit N5
Length=456
Score = 96.7 bits (239), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 45/116 (38%), Positives = 79/116 (68%), Gaps = 0/116 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
+++QVET+G+M+++EK ++IL+QM L + D+IR QI+SKK++ K + + LK+KY
Sbjct 167 QELQVETYGSMEKKEKAEFILEQMRLCIAVKDYIRTQIISKKINTKFFQEEGTEELKLKY 226
Query 62 YELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
Y LM LHE + L + K + IY+T I +D ++ + L+ ++++LAP+D+E
Sbjct 227 YNLMIQVDLHEGSYLSICKHYRAIYDTPCILEDSSKWQQALKSVVLYVILAPYDNE 282
> pfa:PF10_0174 26S proteasome subunit p55, putative; K03035 26S
proteasome regulatory subunit N5
Length=467
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 50/116 (43%), Positives = 84/116 (72%), Gaps = 0/116 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
+DVQVETF +M++ +KT+YIL+QM L+L + DFIRC ++S+K++P LL + LK+KY
Sbjct 172 QDVQVETFISMNKRDKTEYILEQMRLVLLRKDFIRCHVISRKINPSLLNTEEFADLKLKY 231
Query 62 YELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
Y M YY++E++ LDVAKC+ ++T I+ +D + ++ + IF++L+P+D +
Sbjct 232 YMYMIQYYINEESYLDVAKCYEERFHTDIVLNDRNLWIDEMKCYIIFLILSPYDEQ 287
> hsa:5718 PSMD12, MGC75406, Rpn5, p55; proteasome (prosome, macropain)
26S subunit, non-ATPase, 12; K03035 26S proteasome
regulatory subunit N5
Length=456
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 44/116 (37%), Positives = 78/116 (67%), Gaps = 0/116 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
+++QVET+G+M+++E+ ++IL+QM L L D+IR QI+SKK++ K + + + LK+KY
Sbjct 167 QELQVETYGSMEKKERVEFILEQMRLCLAVKDYIRTQIISKKINTKFFQEENTEKLKLKY 226
Query 62 YELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
Y LM HE + L + K + IY+T I+ + + + L+ ++++LAPFD+E
Sbjct 227 YNLMIQLDQHEGSYLSICKHYRAIYDTPCIQAESEKWQQALKSVVLYVILAPFDNE 282
> mmu:66997 Psmd12, 1500002F15Rik, AI480719, P55; proteasome (prosome,
macropain) 26S subunit, non-ATPase, 12; K03035 26S
proteasome regulatory subunit N5
Length=456
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 44/116 (37%), Positives = 78/116 (67%), Gaps = 0/116 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
+++QVET+G+M+++E+ ++IL+QM L L D+IR QI+SKK++ K + + + LK+KY
Sbjct 167 QELQVETYGSMEKKERVEFILEQMRLCLAVKDYIRTQIISKKINTKFFQEENTENLKLKY 226
Query 62 YELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
Y LM HE + L + K + IY+T I+ + + + L+ ++++LAPFD+E
Sbjct 227 YNLMIQLDQHEGSYLSICKHYRAIYDTPCIQAESDKWQQALKSVVLYVILAPFDNE 282
> xla:431923 hypothetical protein MGC81129; K03035 26S proteasome
regulatory subunit N5
Length=441
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 74/116 (63%), Gaps = 0/116 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
+++QVET+G+M+++EK ++IL+QM L L D IR QI+SKK++ K + + + K+KY
Sbjct 152 QELQVETYGSMEKKEKVEFILEQMRLCLAVKDHIRTQIISKKINTKFFQEENTEKPKLKY 211
Query 62 YELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
Y LM HE + L + K + IY T I+ + + L+ ++++L+P+D+E
Sbjct 212 YNLMIQLDQHEGSYLSICKHYRAIYYTPCIQAESDKWQHALKSVVLYIILSPYDNE 267
> xla:399245 psmd12, MGC80339; proteasome (prosome, macropain)
26S subunit, non-ATPase, 12; K03035 26S proteasome regulatory
subunit N5
Length=441
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 73/116 (62%), Gaps = 0/116 (0%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
+++QVET+G+M ++EK ++IL+QM L L DFIR QI+SKK++ K + + + K+ Y
Sbjct 152 QELQVETYGSMVKKEKVEFILEQMRLCLAVKDFIRTQIISKKINTKFFQEENTEKSKLNY 211
Query 62 YELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
Y LM HE + L + K + IY+T I + + L+ ++++L+P+D+E
Sbjct 212 YNLMIQVDQHEGSYLSICKHYRAIYDTPCILAESDKWQHALKSVVLYVILSPYDNE 267
> cel:F10G7.8 rpn-5; proteasome Regulatory Particle, Non-ATPase-like
family member (rpn-5); K03035 26S proteasome regulatory
subunit N5
Length=490
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 45/118 (38%), Positives = 72/118 (61%), Gaps = 3/118 (2%)
Query 3 DVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAAD---LQVLKV 59
++QVET+G+M+ EK +Y+L+QM L ++DF+R I+SKK++ K +D +Q LK+
Sbjct 190 ELQVETYGSMEMREKVQYLLEQMRYSLVRNDFVRATIISKKINIKFFNKSDEDEVQNLKL 249
Query 60 KYYELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
KYY+ M LH+ LDV + IY T+ I+ D A+ L ++ +LAP +E
Sbjct 250 KYYDSMIRIGLHDGNYLDVCRHHREIYETKKIKADSAKATSHLRSAIVYCLLAPHTNE 307
> ath:AT5G64760 RPN5B; RPN5B (REGULATORY PARTICLE NON-ATPASE SUBUNIT
5B); K03035 26S proteasome regulatory subunit N5
Length=442
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 47/137 (34%), Positives = 75/137 (54%), Gaps = 23/137 (16%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEA---------- 51
++V VETFGAM + EK +IL+Q+ L L + DF+R QI+S+K++P++ +A
Sbjct 134 QEVAVETFGAMAKTEKIAFILEQVRLCLDQKDFVRAQILSRKINPRVFDADTTKEKKKPK 193
Query 52 ----------AD---LQVLKVKYYELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARM 98
AD L VLK YYELM YY H + +++ + + IY+ ++++P +
Sbjct 194 EGENMVEEAPADIPTLLVLKRIYYELMIRYYSHNNEYIEICRSYKAIYDIPSVKENPEQW 253
Query 99 NECLEFFSIFMVLAPFD 115
L F+ LAP D
Sbjct 254 IPVLRKICWFLALAPHD 270
> ath:AT5G09900 EMB2107 (EMBRYO DEFECTIVE 2107); K03035 26S proteasome
regulatory subunit N5
Length=462
Score = 81.3 bits (199), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 46/137 (33%), Positives = 74/137 (54%), Gaps = 23/137 (16%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEA---------- 51
++V VETFGAM + EK +IL+Q+ L L + DF+R QI+S+K++P++ +A
Sbjct 134 QEVAVETFGAMAKTEKIAFILEQVRLCLDRQDFVRAQILSRKINPRVFDADTKKDKKKPK 193
Query 52 ----------ADLQV---LKVKYYELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARM 98
AD+ LK YYELM YY H + +++ + + IY+ +++ P +
Sbjct 194 EGDNMVEEAPADIPTLLELKRIYYELMIRYYSHNNEYIEICRSYKAIYDIPSVKETPEQW 253
Query 99 NECLEFFSIFMVLAPFD 115
L F+VLAP D
Sbjct 254 IPVLRKICWFLVLAPHD 270
> sce:YDL147W RPN5, NAS5; Essential, non-ATPase regulatory subunit
of the 26S proteasome lid, similar to mammalian p55 subunit
and to another S. cerevisiae regulatory subunit, Rpn7p;
K03035 26S proteasome regulatory subunit N5
Length=445
Score = 78.2 bits (191), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 68/114 (59%), Gaps = 0/114 (0%)
Query 3 DVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKYY 62
++QVET+G+M+ EK ++IL+QM L + K D+ + ++S+K+ K + + LK++YY
Sbjct 162 ELQVETYGSMEMSEKIQFILEQMELSILKGDYSQATVLSRKILKKTFKNPKYESLKLEYY 221
Query 63 ELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDS 116
L+ LH+ L+VA+ IY T I+ D A+ L F+VL+P+ +
Sbjct 222 NLLVKISLHKREYLEVAQYLQEIYQTDAIKSDEAKWKPVLSHIVYFLVLSPYGN 275
> cpv:cgd1_1530 Rpn5 like 26S proteasomal regulatory subunit 12,
PINT domain containing protein ; K03035 26S proteasome regulatory
subunit N5
Length=559
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 44/146 (30%), Positives = 78/146 (53%), Gaps = 33/146 (22%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEA-ADLQVLKVK 60
ED+ VET G MD EKT+Y+L+QM L L DF+R QI +KK++PK++E DL+V+
Sbjct 217 EDITVETIGNMDLREKTQYVLEQMRLSLLCKDFVRLQIFAKKINPKIIEKFIDLKVI--- 273
Query 61 YYELMTVYYLHEDALLDVAKCFGHIYNT-------------QIIRDDPARMN-------- 99
YY+ + + + +E + +++ CF ++ N+ ++I + P M
Sbjct 274 YYQYLIILWHYEQSPKEISMCFLNLLNSIANFENDTENSYEKLIGEIPEYMKSPISQYNF 333
Query 100 --------ECLEFFSIFMVLAPFDSE 117
C+E + I+++L P+ +
Sbjct 334 SEKMPTTASCIEGYVIYLILGPYSTN 359
> bbo:BBOV_III011560 17.m07987; PCI domain containing protein;
K03035 26S proteasome regulatory subunit N5
Length=474
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 65/115 (56%), Gaps = 1/115 (0%)
Query 3 DVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKYY 62
+ +VETFG + ++EK +Y+L+QM L L +D++R I S K+ ++L+ + K+ YY
Sbjct 178 NTEVETFGILPKKEKVRYLLEQMRLHLLNNDYLRFYIASNKIDDRVLDNDGFEEHKMTYY 237
Query 63 ELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSIFMVLAPFDSE 117
E M Y+LH +VAK + + I D +++ LE IF++++ E
Sbjct 238 EYMVHYHLHSKDYFEVAKAYRQRLDCTIKLDLNDWLSD-LESVVIFLMISAISEE 291
> tpv:TP02_0768 26S proteasome subunit p55; K03035 26S proteasome
regulatory subunit N5
Length=500
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 31/88 (35%), Positives = 57/88 (64%), Gaps = 2/88 (2%)
Query 2 EDVQVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKY 61
++++VET+G++++ EK +YIL+QM + L D+IR + SKK++ L+ D K+++
Sbjct 176 QNIEVETYGSLNKLEKIRYILEQMRVNLLNGDYIRFFMTSKKITESALD--DYVPEKLQF 233
Query 62 YELMTVYYLHEDALLDVAKCFGHIYNTQ 89
Y+ M YY H+ + +V K IY+T+
Sbjct 234 YDFMIQYYHHDFDIENVTKSLYTIYSTK 261
> dre:565026 tgfbrap1, MGC162302, zgc:162302; transforming growth
factor, beta receptor associated protein 1
Length=863
Score = 29.3 bits (64), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 33/73 (45%), Gaps = 13/73 (17%)
Query 26 MLLLRKSDFIRCQIVSKKLSPKLLEAADLQVL------KVKYYELMTVYYLHEDALLDVA 79
+LL S F RC P L E ADL L KV+ ++ + YLHE D+A
Sbjct 389 LLLPASSSFTRCH-------PPLHEFADLNHLTQGDQEKVQRFKRFLISYLHEVRSSDIA 441
Query 80 KCFGHIYNTQIIR 92
F +T +++
Sbjct 442 NGFHEDVDTALLK 454
> sce:YMR282C AEP2, ATP13; Aep2p
Length=580
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 24/80 (30%), Positives = 39/80 (48%), Gaps = 8/80 (10%)
Query 13 DREEKTKYILKQMMLLLRKSDFIRCQIVSKKLSPK-LLEAADLQVLKVKYYELMTVYYLH 71
D E+ + L++M+ L K+ IR K + + LL+ ADL + + K
Sbjct 264 DNSEQKQGFLRKMVRLGAKNTSIRLSSTYKAMDHQTLLKIADLALQEKKLLN-------S 316
Query 72 EDALLDVAKCFGHIYNTQII 91
ED L + + FGH+ TQI+
Sbjct 317 EDLLSTLIQSFGHLGQTQIL 336
> eco:b1285 gmr, ECK1280, JW1278, yciR; cyclic-di-GMP phosphodiesterase;
CsgD regulator; modulator of RNase II stability;
K14051 cyclic di-GMP phosphodiesterase Gmr [EC:3.1.4.52]
Length=661
Score = 29.3 bits (64), Expect = 3.6, Method: Composition-based stats.
Identities = 18/59 (30%), Positives = 27/59 (45%), Gaps = 8/59 (13%)
Query 67 VYYLHEDALLDVAKCFGHIYNTQIIRDDPARMNECLEFFSI--------FMVLAPFDSE 117
V YL D V +GH++ Q++RD + CLE + F+VLA S+
Sbjct 269 VVYLDLDNFKKVNDAYGHLFGDQLLRDVSLAILSCLEHDQVLARPGGDEFLVLASNTSQ 327
> dre:100170827 vars2, si:ch211-152p11.2; valyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.9); K01873 valyl-tRNA
synthetase [EC:6.1.1.9]
Length=1057
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 26/38 (68%), Gaps = 2/38 (5%)
Query 28 LLRKSDFIRCQIVSKKLSPKLLEAADLQVLKVKYYELM 65
LL+K F+RCQ ++KK + + +E DLQ++ ++Y M
Sbjct 461 LLKKQWFVRCQEMAKK-AVQAVEEGDLQIIP-QFYSKM 496
> dre:561122 nlgn4b, nlgn4a; neuroligin 4b; K07378 neuroligin
Length=826
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 21/76 (27%), Positives = 43/76 (56%), Gaps = 4/76 (5%)
Query 38 QIVSKKLSPKLLEAADL-QVLKVKYYELMTVYYLHEDALLDVAKCFGHIYNTQIIRDDPA 96
+I+++K+ +L++ DL + L+ K Y+ + Y+ + A +A FG + + +I DDP
Sbjct 295 RILAEKVGCNMLDSIDLVECLQNKNYKELIEQYITQ-AKYHIA--FGPVIDGDVIPDDPQ 351
Query 97 RMNECLEFFSIFMVLA 112
+ E EF + ++L
Sbjct 352 ILMEQGEFLNYDIMLG 367
> cel:F59A6.5 hypothetical protein
Length=1286
Score = 27.7 bits (60), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 5 QVETFGAMDREEKTKYILKQMMLLLRKSDFIRCQIV 40
Q T D E+ TKY+ ++ M L R+ IRC+++
Sbjct 313 QTTTASLGDSEQATKYLHEENMKLTRQKADIRCELL 348
> cel:Y110A7A.19 hypothetical protein
Length=660
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 16/59 (27%), Positives = 31/59 (52%), Gaps = 2/59 (3%)
Query 3 DVQVETFGAMDREEKTKYILKQMMLLLRKSDFI--RCQIVSKKLSPKLLEAADLQVLKV 59
+VQ+ T G +RE+ T + K + + + S F R + + +KLSP + L + ++
Sbjct 503 NVQLHTLGTSEREQFTSLVQKLVAVWVEFSQFTEERMRRLQRKLSPSQISECALLLTRI 561
Lambda K H
0.326 0.140 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2032807080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40