bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5457_orf1
Length=148
Score E
Sequences producing significant alignments: (Bits) Value
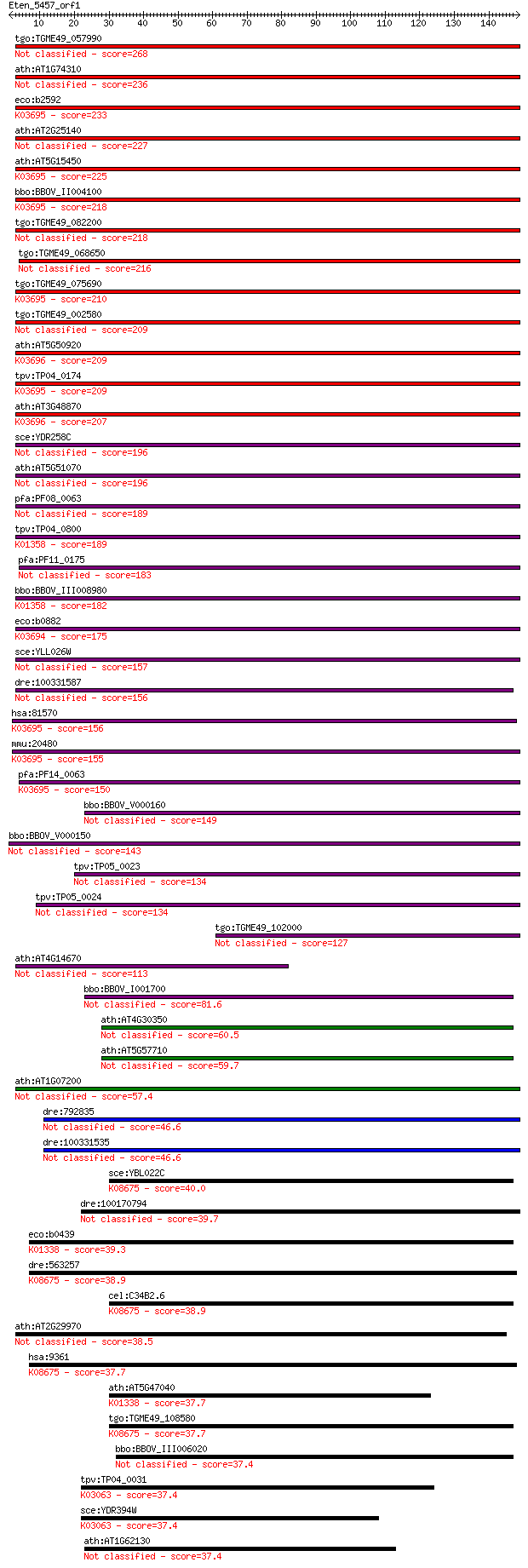
tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53) 268 6e-72
ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEI... 236 3e-62
eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation ... 233 1e-61
ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP b... 227 1e-59
ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP b... 225 4e-59
bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp pr... 218 6e-57
tgo:TGME49_082200 clpB protein, putative 218 6e-57
tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.2... 216 2e-56
tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.2... 210 1e-54
tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53) 209 2e-54
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 209 2e-54
tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp p... 209 3e-54
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 207 7e-54
sce:YDR258C HSP78; Hsp78p 196 2e-50
ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ... 196 2e-50
pfa:PF08_0063 ClpB protein, putative 189 3e-48
tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit (... 189 3e-48
pfa:PF11_0175 heat shock protein 101, putative 183 2e-46
bbo:BBOV_III008980 17.m07783; Clp amino terminal domain contai... 182 2e-46
eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity ... 175 6e-44
sce:YLL026W HSP104; Heat shock protein that cooperates with Yd... 157 1e-38
dre:100331587 suppressor of K+ transport defect 3-like 156 2e-38
hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic pepti... 156 3e-38
mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase B ... 155 4e-38
pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP... 150 1e-36
bbo:BBOV_V000160 clpC 149 4e-36
bbo:BBOV_V000150 clpC 143 2e-34
tpv:TP05_0023 clpC; molecular chaperone 134 9e-32
tpv:TP05_0024 clpC; molecular chaperone 134 9e-32
tgo:TGME49_102000 chaperone clpB protein, putative 127 1e-29
ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosph... 113 2e-25
bbo:BBOV_I001700 19.m02115; chaperone clpB 81.6 7e-16
ath:AT4G30350 heat shock protein-related 60.5 2e-09
ath:AT5G57710 heat shock protein-related 59.7 3e-09
ath:AT1G07200 ATP-dependent Clp protease ClpB protein-related 57.4 2e-08
dre:792835 torsin family 3, member A 46.6 3e-05
dre:100331535 torsin family 3, member A-like 46.6 3e-05
sce:YBL022C PIM1; ATP-dependent Lon protease, involved in degr... 40.0 0.003
dre:100170794 zgc:194342 39.7 0.004
eco:b0439 lon, capR, deg, dir, ECK0433, JW0429, lonA, lopA, mu... 39.3 0.005
dre:563257 lonp1, fc64d11, prss15, wu:fc64d11; lon peptidase 1... 38.9 0.007
cel:C34B2.6 hypothetical protein; K08675 Lon-like ATP-dependen... 38.9 0.007
ath:AT2G29970 heat shock protein-related 38.5 0.007
hsa:9361 LONP1, LON, LONP, LonHS, MGC1498, PIM1, PRSS15, hLON;... 37.7 0.012
ath:AT5G47040 LON2; LON2 (LON PROTEASE 2); ATP binding / ATP-d... 37.7 0.013
tgo:TGME49_108580 lon protease, putative (EC:3.4.21.53); K0867... 37.7 0.014
bbo:BBOV_III006020 17.m07531; ATP-dependent protease La family... 37.4 0.015
tpv:TP04_0031 26S proteasome aaa-ATPase subunit Rpt3 (EC:3.6.4... 37.4 0.016
sce:YDR394W RPT3, YNT1, YTA2; One of six ATPases of the 19S re... 37.4 0.019
ath:AT1G62130 AAA-type ATPase family protein 37.4 0.019
> tgo:TGME49_057990 heat shock protein, putative (EC:3.4.21.53)
Length=921
Score = 268 bits (684), Expect = 6e-72, Method: Compositional matrix adjust.
Identities = 123/146 (84%), Positives = 137/146 (93%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV AV+++ILRSAAGL+RRN+PIGSFLFLGPTGVGKTELCK VAE LFD+KER+VR DMS
Sbjct 578 AVEAVTQAILRSAAGLSRRNRPIGSFLFLGPTGVGKTELCKRVAESLFDSKERLVRFDMS 637
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HSV+RLIGAPPGYVGH+EGGQLTE +RRNPYSVVL DEVEKAH QVWNVLLQVLD
Sbjct 638 EYMEQHSVSRLIGAPPGYVGHDEGGQLTEEIRRNPYSVVLFDEVEKAHSQVWNVLLQVLD 697
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTDSQGRTVDFSN ++ILTSN+G
Sbjct 698 DGRLTDSQGRTVDFSNTIIILTSNLG 723
> ath:AT1G74310 ATHSP101 (ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN
101); ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=911
Score = 236 bits (601), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 111/146 (76%), Positives = 126/146 (86%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV AVSE+ILRS AGL R +P GSFLFLGPTGVGKTEL KA+AE+LFD + +VR+DMS
Sbjct 577 AVNAVSEAILRSRAGLGRPQQPTGSFLFLGPTGVGKTELAKALAEQLFDDENLLVRIDMS 636
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HSV+RLIGAPPGYVGHEEGGQLTEAVRR PY V+L DEVEKAH V+N LLQVLD
Sbjct 637 EYMEQHSVSRLIGAPPGYVGHEEGGQLTEAVRRRPYCVILFDEVEKAHVAVFNTLLQVLD 696
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTD QGRTVDF N ++I+TSN+G
Sbjct 697 DGRLTDGQGRTVDFRNSVIIMTSNLG 722
> eco:b2592 clpB, ECK2590, htpM, JW2573; protein disaggregation
chaperone; K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=857
Score = 233 bits (595), Expect = 1e-61, Method: Compositional matrix adjust.
Identities = 109/146 (74%), Positives = 128/146 (87%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV AVS +I RS AGLA N+PIGSFLFLGPTGVGKTELCKA+A +FD+ E +VR+DMS
Sbjct 576 AVDAVSNAIRRSRAGLADPNRPIGSFLFLGPTGVGKTELCKALANFMFDSDEAMVRIDMS 635
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ME HSV+RL+GAPPGYVG+EEGG LTEAVRR PYSV+LLDEVEKAH V+N+LLQVLD
Sbjct 636 EFMEKHSVSRLVGAPPGYVGYEEGGYLTEAVRRRPYSVILLDEVEKAHPDVFNILLQVLD 695
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTD QGRTVDF N ++I+TSN+G
Sbjct 696 DGRLTDGQGRTVDFRNTVVIMTSNLG 721
> ath:AT2G25140 CLPB4; CLPB4 (CASEIN LYTIC PROTEINASE B4); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding
Length=964
Score = 227 bits (578), Expect = 1e-59, Method: Compositional matrix adjust.
Identities = 103/146 (70%), Positives = 127/146 (86%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV +V+++I RS AGL+ N+PI SF+F+GPTGVGKTEL KA+A LF+T+ IVR+DMS
Sbjct 661 AVKSVADAIRRSRAGLSDPNRPIASFMFMGPTGVGKTELAKALAGYLFNTENAIVRVDMS 720
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HSV+RL+GAPPGYVG+EEGGQLTE VRR PYSVVL DE+EKAH V+N+LLQ+LD
Sbjct 721 EYMEKHSVSRLVGAPPGYVGYEEGGQLTEVVRRRPYSVVLFDEIEKAHPDVFNILLQLLD 780
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GR+TDSQGRTV F NC++I+TSNIG
Sbjct 781 DGRITDSQGRTVSFKNCVVIMTSNIG 806
> ath:AT5G15450 CLPB3; CLPB3 (CASEIN LYTIC PROTEINASE B3); ATP
binding / ATPase/ nucleoside-triphosphatase/ nucleotide binding
/ protein binding; K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=968
Score = 225 bits (574), Expect = 4e-59, Method: Compositional matrix adjust.
Identities = 101/146 (69%), Positives = 127/146 (86%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AVTAV+E+I RS AGL+ +PI SF+F+GPTGVGKTEL KA+A +F+T+E +VR+DMS
Sbjct 656 AVTAVAEAIQRSRAGLSDPGRPIASFMFMGPTGVGKTELAKALASYMFNTEEALVRIDMS 715
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME H+V+RLIGAPPGYVG+EEGGQLTE VRR PYSV+L DE+EKAH V+NV LQ+LD
Sbjct 716 EYMEKHAVSRLIGAPPGYVGYEEGGQLTETVRRRPYSVILFDEIEKAHGDVFNVFLQILD 775
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GR+TDSQGRTV F+N ++I+TSN+G
Sbjct 776 DGRVTDSQGRTVSFTNTVIIMTSNVG 801
> bbo:BBOV_II004100 18.m06340; ClpB; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=931
Score = 218 bits (555), Expect = 6e-57, Method: Compositional matrix adjust.
Identities = 94/146 (64%), Positives = 121/146 (82%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV V+ ++ RS G+ +PI +FLGPTGVGKTELCKA+AE+LFDT E I+R DMS
Sbjct 647 AVDIVTRAVQRSRVGMNDPKRPIAGLMFLGPTGVGKTELCKAIAEQLFDTDEAIIRFDMS 706
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HSV+RL+GAPPGY+G+++GG LTEAVRR PYS+VL DE+EKAH V+N++LQ+LD
Sbjct 707 EYMEKHSVSRLVGAPPGYIGYDQGGLLTEAVRRRPYSIVLFDEIEKAHPDVFNIMLQLLD 766
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTDS GR V+F+NC++I TSN+G
Sbjct 767 DGRLTDSSGRKVNFTNCMIIFTSNLG 792
> tgo:TGME49_082200 clpB protein, putative
Length=970
Score = 218 bits (555), Expect = 6e-57, Method: Compositional matrix adjust.
Identities = 95/146 (65%), Positives = 127/146 (86%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV AV+++++R+ AGL+R P+GSFLFLGPTGVGKTEL KA+A E+F +++ ++R+DMS
Sbjct 593 AVKAVADAMVRARAGLSREGMPVGSFLFLGPTGVGKTELAKALAMEMFHSEKNLIRIDMS 652
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ E+HSV+RLIG+PPGYVGH+ GGQLTEAVRR P+SVVL DE+EK H Q+ N++LQ+LD
Sbjct 653 EFSEAHSVSRLIGSPPGYVGHDAGGQLTEAVRRRPHSVVLFDEIEKGHQQILNIMLQMLD 712
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
EGRLTD +G VDF+NC++ILTSN+G
Sbjct 713 EGRLTDGKGLLVDFTNCVIILTSNVG 738
> tgo:TGME49_068650 clp ATP-binding chain B1, putative (EC:3.4.21.53)
Length=929
Score = 216 bits (550), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 97/145 (66%), Positives = 121/145 (83%), Gaps = 0/145 (0%)
Query 4 VTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSE 63
V +V+E+I RS AGL N+PI S +FLGPTGVGKTELCKA+A +LFDT+E ++R DMSE
Sbjct 603 VRSVAEAIQRSRAGLCDPNRPIASLVFLGPTGVGKTELCKALARQLFDTEEALLRFDMSE 662
Query 64 YMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDE 123
YME HS ARLIGAPPGY+G ++GGQLTEAVRR PYSV+L DE+EKAH +V+N+ LQ+L++
Sbjct 663 YMEKHSTARLIGAPPGYIGFDKGGQLTEAVRRRPYSVLLFDEMEKAHPEVFNIFLQILED 722
Query 124 GRLTDSQGRTVDFSNCLLILTSNIG 148
G LTDS G TV F NC++I TSN+G
Sbjct 723 GILTDSHGHTVSFKNCIIIFTSNMG 747
> tgo:TGME49_075690 chaperone clpB 1 protein, putative (EC:3.4.21.53);
K03695 ATP-dependent Clp protease ATP-binding subunit
ClpB
Length=898
Score = 210 bits (535), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 91/146 (62%), Positives = 123/146 (84%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV V+E+I RS AGL N+PI S FLGPTGVGKTELC+++AE +FD+++ +V++DMS
Sbjct 734 AVQVVAEAIQRSRAGLNDPNRPIASLFFLGPTGVGKTELCRSLAELMFDSEDAMVKIDMS 793
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME H+++RL+GAPPGYVG+E+GGQLT+ VR+ PYSV+L DE+EKAH V+NVLLQ+LD
Sbjct 794 EYMEKHTISRLLGAPPGYVGYEQGGQLTDEVRKKPYSVILFDEMEKAHPDVFNVLLQILD 853
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GR+TD +G V+F NC++I TSN+G
Sbjct 854 DGRVTDGKGNVVNFRNCIVIFTSNLG 879
> tgo:TGME49_002580 heat shock protein, putative (EC:3.4.21.53)
Length=983
Score = 209 bits (533), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 98/146 (67%), Positives = 121/146 (82%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV AV+ +I+RS AGLARRN PIG+FLFLG TGVGKTEL KA+ +F ++ ++RLDMS
Sbjct 606 AVEAVANAIIRSRAGLARRNAPIGTFLFLGSTGVGKTELAKALTAAIFHDEKNLIRLDMS 665
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EY E H+VARL+G+PPGYV H+EGGQLTEAVR+ P+SVVL DE+E AH V+ LLQ+LD
Sbjct 666 EYSEPHTVARLVGSPPGYVSHDEGGQLTEAVRQRPHSVVLFDEIENAHPNVFAYLLQLLD 725
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
EGRLTD +G TVDF+NC++I TSNIG
Sbjct 726 EGRLTDMRGITVDFTNCVIIATSNIG 751
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 209 bits (533), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 94/146 (64%), Positives = 122/146 (83%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV A+S +I R+ GL N+PI SF+F GPTGVGK+EL KA+A F ++E ++RLDMS
Sbjct 616 AVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMS 675
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ME H+V++LIG+PPGYVG+ EGGQLTEAVRR PY+VVL DE+EKAH V+N++LQ+L+
Sbjct 676 EFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILE 735
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTDS+GRTVDF N LLI+TSN+G
Sbjct 736 DGRLTDSKGRTVDFKNTLLIMTSNVG 761
> tpv:TP04_0174 hypothetical protein; K03695 ATP-dependent Clp
protease ATP-binding subunit ClpB
Length=985
Score = 209 bits (532), Expect = 3e-54, Method: Composition-based stats.
Identities = 93/146 (63%), Positives = 121/146 (82%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
A+ AV ++ RS G+ KPI + +FLGPTGVGKTEL KA+AE+LFD++E I+R DMS
Sbjct 702 AIDAVVNAVQRSRVGMNDPKKPIAALMFLGPTGVGKTELSKAIAEQLFDSEEAIIRFDMS 761
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HSV++L+GAPPGY+G+E+GG LTEA+RR PYS++L DE+EKAH V+N+LLQVLD
Sbjct 762 EYMEKHSVSKLVGAPPGYIGYEQGGLLTEAIRRKPYSILLFDEIEKAHSDVYNILLQVLD 821
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTDS GR V+F+N L+I TSN+G
Sbjct 822 DGRLTDSLGRKVNFTNSLIIFTSNLG 847
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 207 bits (528), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 93/146 (63%), Positives = 122/146 (83%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV A+S +I R+ GL N+PI SF+F GPTGVGK+EL KA+A F ++E ++RLDMS
Sbjct 637 AVKAISRAIRRARVGLKNPNRPIASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMS 696
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ME H+V++LIG+PPGYVG+ EGGQLTEAVRR PY++VL DE+EKAH V+N++LQ+L+
Sbjct 697 EFMERHTVSKLIGSPPGYVGYTEGGQLTEAVRRRPYTLVLFDEIEKAHPDVFNMMLQILE 756
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GRLTDS+GRTVDF N LLI+TSN+G
Sbjct 757 DGRLTDSKGRTVDFKNTLLIMTSNVG 782
> sce:YDR258C HSP78; Hsp78p
Length=811
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 91/146 (62%), Positives = 112/146 (76%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
A+ A+S+++ AGL +PI SF+FLGPTG GKTEL KA+AE LFD + ++R DMS
Sbjct 512 AIAAISDAVRLQRAGLTSEKRPIASFMFLGPTGTGKTELTKALAEFLFDDESNVIRFDMS 571
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E+ E H+V+RLIGAPPGYV E GGQLTEAVRR PY+VVL DE EKAH V +LLQVLD
Sbjct 572 EFQEKHTVSRLIGAPPGYVLSESGGQLTEAVRRKPYAVVLFDEFEKAHPDVSKLLLQVLD 631
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
EG+LTDS G VDF N ++++TSNIG
Sbjct 632 EGKLTDSLGHHVDFRNTIIVMTSNIG 657
> ath:AT5G51070 ERD1; ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1);
ATP binding / ATPase/ nucleoside-triphosphatase/ nucleotide
binding / protein binding
Length=945
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 88/146 (60%), Positives = 116/146 (79%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV A+S ++ RS GL ++PI + LF GPTGVGKTEL KA+A F ++E ++RLDMS
Sbjct 635 AVAAISRAVKRSRVGLKDPDRPIAAMLFCGPTGVGKTELTKALAANYFGSEESMLRLDMS 694
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME H+V++LIG+PPGYVG EEGG LTEA+RR P++VVL DE+EKAH ++N+LLQ+ +
Sbjct 695 EYMERHTVSKLIGSPPGYVGFEEGGMLTEAIRRRPFTVVLFDEIEKAHPDIFNILLQLFE 754
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+G LTDSQGR V F N L+I+TSN+G
Sbjct 755 DGHLTDSQGRRVSFKNALIIMTSNVG 780
> pfa:PF08_0063 ClpB protein, putative
Length=1070
Score = 189 bits (480), Expect = 3e-48, Method: Composition-based stats.
Identities = 82/146 (56%), Positives = 115/146 (78%), Gaps = 0/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV V++++ RS G+ +PI S +FLGPTGVGKTEL K +A+ LFDT E ++ DMS
Sbjct 787 AVKVVTKAVQRSRVGMNNPKRPIASLMFLGPTGVGKTELSKVLADVLFDTPEAVIHFDMS 846
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EYME HS+++LIGA PGYVG+E+GG LT+AVR+ PYS++L DE+EKAH V+N+LL+V+D
Sbjct 847 EYMEKHSISKLIGAAPGYVGYEQGGLLTDAVRKKPYSIILFDEIEKAHPDVYNLLLRVID 906
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
EG+L+D++G +F N ++I TSN+G
Sbjct 907 EGKLSDTKGNVANFRNTIIIFTSNLG 932
> tpv:TP04_0800 ATP-dependent Clp protease ATP-binding subunit
(EC:3.4.21.92); K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=900
Score = 189 bits (480), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 81/146 (55%), Positives = 114/146 (78%), Gaps = 1/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV V ++I R+ + N+PIGSFLF GP GVGK+E+ +A+ + LF KE ++R+DMS
Sbjct 581 AVKNVCKAIRRAKTNIKNPNRPIGSFLFCGPPGVGKSEVARALTKYLF-AKENLIRIDMS 639
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EY E HS++R++G+PPGY GH+ GGQLTE V+ NPYSVV+ DE+EKAHH V N+LLQ+L+
Sbjct 640 EYTEPHSISRILGSPPGYKGHDTGGQLTEKVKSNPYSVVMFDEIEKAHHDVLNILLQILE 699
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+G+LTDS+ +T+ F N ++I+TSN G
Sbjct 700 DGKLTDSKNQTISFKNTIIIMTSNTG 725
> pfa:PF11_0175 heat shock protein 101, putative
Length=906
Score = 183 bits (465), Expect = 2e-46, Method: Composition-based stats.
Identities = 79/145 (54%), Positives = 117/145 (80%), Gaps = 0/145 (0%)
Query 4 VTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSE 63
+ ++S++++++A G+ KPIG+FLFLGPTGVGKTEL K +A ELF++K+ ++R++MSE
Sbjct 610 IKSLSDAVVKAATGMKDPEKPIGTFLFLGPTGVGKTELAKTLAIELFNSKDNLIRVNMSE 669
Query 64 YMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDE 123
+ E+HSV+++ G+PPGYVG + GQLTEAVR P+SVVL DE+EKAH V+ VLLQ+L +
Sbjct 670 FTEAHSVSKITGSPPGYVGFSDSGQLTEAVREKPHSVVLFDELEKAHADVFKVLLQILGD 729
Query 124 GRLTDSQGRTVDFSNCLLILTSNIG 148
G + D+ R +DFSN ++I+TSN+G
Sbjct 730 GYINDNHRRNIDFSNTIIIMTSNLG 754
> bbo:BBOV_III008980 17.m07783; Clp amino terminal domain containing
protein; K01358 ATP-dependent Clp protease, protease
subunit [EC:3.4.21.92]
Length=1005
Score = 182 bits (463), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 77/146 (52%), Positives = 115/146 (78%), Gaps = 1/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV ++++I R+ + +PIGSFLF GP GVGK+E+ K + + +F T++ +++LDMS
Sbjct 667 AVKNIAKAIRRAKTNIKNPERPIGSFLFCGPPGVGKSEIAKTLTKLMF-TEDNLIKLDMS 725
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
EY E HS++R++G+PPGY GH+ GGQLTE +R+NPYSVV+ DE+EKAH V N+LLQ+L+
Sbjct 726 EYNEPHSISRILGSPPGYKGHDSGGQLTEKLRKNPYSVVMFDEIEKAHRNVLNILLQILE 785
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+G+LTDS+ +TV F N ++I+TSN+G
Sbjct 786 DGKLTDSKNQTVSFKNTIIIMTSNVG 811
> eco:b0882 clpA, ECK0873, JW0866, lopD; ATPase and specificity
subunit of ClpA-ClpP ATP-dependent serine protease, chaperone
activity; K03694 ATP-dependent Clp protease ATP-binding
subunit ClpA
Length=758
Score = 175 bits (443), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 85/150 (56%), Positives = 113/150 (75%), Gaps = 11/150 (7%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTE----LCKAVAEELFDTKERIVR 58
A+ A++E+I + AGL +KP+GSFLF GPTGVGKTE L KA+ EL +R
Sbjct 466 AIEALTEAIKMARAGLGHEHKPVGSFLFAGPTGVGKTEVTVQLSKALGIEL-------LR 518
Query 59 LDMSEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLL 118
DMSEYME H+V+RLIGAPPGYVG ++GG LT+AV ++P++V+LLDE+EKAH V+N+LL
Sbjct 519 FDMSEYMERHTVSRLIGAPPGYVGFDQGGLLTDAVIKHPHAVLLLDEIEKAHPDVFNILL 578
Query 119 QVLDEGRLTDSQGRTVDFSNCLLILTSNIG 148
QV+D G LTD+ GR DF N +L++T+N G
Sbjct 579 QVMDNGTLTDNNGRKADFRNVVLVMTTNAG 608
> sce:YLL026W HSP104; Heat shock protein that cooperates with
Ydj1p (Hsp40) and Ssa1p (Hsp70) to refold and reactivate previously
denatured, aggregated proteins; responsive to stresses
including: heat, ethanol, and sodium arsenite; involved in
[PSI+] propagation
Length=908
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 76/146 (52%), Positives = 108/146 (73%), Gaps = 1/146 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
A+ AVS ++ S +GLA +P SFLFLG +G GKTEL K VA LF+ ++ ++R+D S
Sbjct 586 AIKAVSNAVRLSRSGLANPRQP-ASFLFLGLSGSGKTELAKKVAGFLFNDEDMMIRVDCS 644
Query 63 EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLD 122
E E ++V++L+G GYVG++EGG LT ++ PYSV+L DEVEKAH V V+LQ+LD
Sbjct 645 ELSEKYAVSKLLGTTAGYVGYDEGGFLTNQLQYKPYSVLLFDEVEKAHPDVLTVMLQMLD 704
Query 123 EGRLTDSQGRTVDFSNCLLILTSNIG 148
+GR+T QG+T+D SNC++I+TSN+G
Sbjct 705 DGRITSGQGKTIDCSNCIVIMTSNLG 730
> dre:100331587 suppressor of K+ transport defect 3-like
Length=409
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 74/145 (51%), Positives = 103/145 (71%), Gaps = 2/145 (1%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELF-DTKERIVRLDM 61
A+ V+ +I R G P+ FLFLG +G+GKTEL K VA + D K+ +R+DM
Sbjct 64 AINTVASAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQVARYMHKDIKKGFIRMDM 122
Query 62 SEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVL 121
SE+ E H VA+ IG+PPGYVGH+EGGQLT+ ++++P +VVL DEVEKAH V V+LQ+
Sbjct 123 SEFQEKHEVAKFIGSPPGYVGHDEGGQLTKQLKQSPSAVVLFDEVEKAHPDVLTVMLQLF 182
Query 122 DEGRLTDSQGRTVDFSNCLLILTSN 146
DEGRLTD +G+T++ + + I+TSN
Sbjct 183 DEGRLTDGKGKTIECKDAIFIMTSN 207
> hsa:81570 CLPB, FLJ13152, HSP78, SKD3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease
ATP-binding subunit ClpB
Length=707
Score = 156 bits (394), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 74/147 (50%), Positives = 103/147 (70%), Gaps = 2/147 (1%)
Query 2 AAVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELF-DTKERIVRLD 60
+A+ V +I R G P+ FLFLG +G+GKTEL K A+ + D K+ +RLD
Sbjct 352 SAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKDAKKGFIRLD 410
Query 61 MSEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQV 120
MSE+ E H VA+ IG+PPGYVGHEEGGQLT+ +++ P +VVL DEV+KAH V ++LQ+
Sbjct 411 MSEFQERHEVAKFIGSPPGYVGHEEGGQLTKKLKQCPNAVVLFDEVDKAHPDVLTIMLQL 470
Query 121 LDEGRLTDSQGRTVDFSNCLLILTSNI 147
DEGRLTD +G+T+D + + I+TSN+
Sbjct 471 FDEGRLTDGKGKTIDCKDAIFIMTSNV 497
> mmu:20480 Clpb, AL118244, Skd3; ClpB caseinolytic peptidase
B homolog (E. coli); K03695 ATP-dependent Clp protease ATP-binding
subunit ClpB
Length=677
Score = 155 bits (393), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 73/148 (49%), Positives = 103/148 (69%), Gaps = 2/148 (1%)
Query 2 AAVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELF-DTKERIVRLD 60
+A+ V +I R G P+ FLFLG +G+GKTEL K A+ + D K+ +RLD
Sbjct 322 SAIATVGAAIRRKENGWYDEEHPL-VFLFLGSSGIGKTELAKQTAKYMHKDAKKGFIRLD 380
Query 61 MSEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQV 120
MSE+ E H VA+ IG+PPGY+GHEEGGQLT+ +++ P +VVL DEV+KAH V ++LQ+
Sbjct 381 MSEFQERHEVAKFIGSPPGYIGHEEGGQLTKKLKQCPNAVVLFDEVDKAHPDVLTIMLQL 440
Query 121 LDEGRLTDSQGRTVDFSNCLLILTSNIG 148
DEGRLTD +G+T+D + + I+TSN+
Sbjct 441 FDEGRLTDGKGKTIDCKDAIFIMTSNVA 468
> pfa:PF14_0063 ATP-dependent CLP protease, putative; K03695 ATP-dependent
Clp protease ATP-binding subunit ClpB
Length=1341
Score = 150 bits (379), Expect = 1e-36, Method: Composition-based stats.
Identities = 66/145 (45%), Positives = 105/145 (72%), Gaps = 1/145 (0%)
Query 4 VTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSE 63
+ +S+ + ++ + NKPIG+ L G +GVGKT + +++ LF+ IV ++MSE
Sbjct 940 IDILSKYLFKAITNIKDPNKPIGTLLLCGSSGVGKTLCAQVISKYLFNEDNLIV-INMSE 998
Query 64 YMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDE 123
Y++ HSV++L G+ PGYVG++EGG+LTE+V++ P+S++L DE+EKAH +V +VLLQ+LD
Sbjct 999 YIDKHSVSKLFGSYPGYVGYKEGGELTESVKKKPFSIILFDEIEKAHSEVLHVLLQILDN 1058
Query 124 GRLTDSQGRTVDFSNCLLILTSNIG 148
G LTDS+G V F N + +T+N+G
Sbjct 1059 GLLTDSKGNKVSFKNTFIFMTTNVG 1083
> bbo:BBOV_V000160 clpC
Length=551
Score = 149 bits (375), Expect = 4e-36, Method: Composition-based stats.
Identities = 65/126 (51%), Positives = 89/126 (70%), Gaps = 0/126 (0%)
Query 23 KPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVG 82
KPIGSFL GP+G GKTE+ K + L+ ++ +++ DMSEY ESHSV++LIGAPPGYVG
Sbjct 301 KPIGSFLLCGPSGTGKTEIVKLITNYLYKSQTNLLQFDMSEYKESHSVSKLIGAPPGYVG 360
Query 83 HEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDFSNCLLI 142
HE GG L + +VL DE+EKA ++++ LQ+LDEG LTDS+G + F+ L+
Sbjct 361 HESGGNLINKINSVESPIVLFDEIEKADKNIFSIFLQILDEGLLTDSKGNSCKFNKSLIF 420
Query 143 LTSNIG 148
TSN+G
Sbjct 421 FTSNLG 426
> bbo:BBOV_V000150 clpC
Length=541
Score = 143 bits (361), Expect = 2e-34, Method: Composition-based stats.
Identities = 64/148 (43%), Positives = 101/148 (68%), Gaps = 1/148 (0%)
Query 1 IAAVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLD 60
I + V +SI +S + + KP+ SFLF GP+G GKTEL K LF + +++++L+
Sbjct 255 INLIEPVVKSINKSFY-IPSKTKPLSSFLFCGPSGAGKTELAKIFTYSLFKSTKQLIKLN 313
Query 61 MSEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQV 120
MSEYME+HS+++++G+PPGYVG+ E V+ P V+L DE+EKAH + +++LQ+
Sbjct 314 MSEYMEAHSISKILGSPPGYVGYNENNDFINKVKSMPNCVILFDEIEKAHKSINDLMLQL 373
Query 121 LDEGRLTDSQGRTVDFSNCLLILTSNIG 148
L+EG+LT + G + F+N +I TSN+G
Sbjct 374 LEEGKLTAANGDVLTFNNSFIIFTSNLG 401
> tpv:TP05_0023 clpC; molecular chaperone
Length=502
Score = 134 bits (338), Expect = 9e-32, Method: Composition-based stats.
Identities = 62/132 (46%), Positives = 91/132 (68%), Gaps = 3/132 (2%)
Query 20 RRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPG 79
+ NKPI +FL GP+G GKT++CK ++ L K+ +++LDMSE E HSV+RL+G+PPG
Sbjct 253 KYNKPIANFLLCGPSGTGKTDICKILSTYLGSEKQNLIKLDMSELAEEHSVSRLLGSPPG 312
Query 80 YVG---HEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDF 136
YVG ++ L + + P SVVLLDE+EKA+ ++ + LQ+LDEG L DS G +F
Sbjct 313 YVGGGKSKKSKTLVDEIIDKPNSVVLLDEIEKAYKRLCYIFLQILDEGILIDSSGTVGNF 372
Query 137 SNCLLILTSNIG 148
+N ++ TSN+G
Sbjct 373 TNSFVVFTSNLG 384
> tpv:TP05_0024 clpC; molecular chaperone
Length=529
Score = 134 bits (337), Expect = 9e-32, Method: Composition-based stats.
Identities = 61/144 (42%), Positives = 98/144 (68%), Gaps = 4/144 (2%)
Query 9 ESILRSAAGLARRN----KPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEY 64
ES+L L+ N KP+GS+L GP+G GKTEL K + +F++ + +++++M+EY
Sbjct 251 ESVLYRIKKLSNANIDETKPLGSWLLCGPSGTGKTELAKILCYTIFNSHDNLIKINMAEY 310
Query 65 MESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEG 124
+E H+V++LIG+PPGY G+ E L+ + V+L DE+EKAH + +++LQ+LD+G
Sbjct 311 VEKHAVSKLIGSPPGYSGYGEDTILSTKFKTGSSFVILFDEIEKAHTSITDLMLQILDKG 370
Query 125 RLTDSQGRTVDFSNCLLILTSNIG 148
+LT S G ++F+N +I TSNIG
Sbjct 371 KLTLSNGDIINFNNSFIIFTSNIG 394
> tgo:TGME49_102000 chaperone clpB protein, putative
Length=240
Score = 127 bits (320), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 52/88 (59%), Positives = 73/88 (82%), Gaps = 0/88 (0%)
Query 61 MSEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQV 120
MSE+ME HS+++LIGAPPGYVG+ + G LTEA+ + P++V+L DE+EKAH + N++LQ+
Sbjct 1 MSEFMEKHSLSKLIGAPPGYVGYGKSGLLTEAIAKKPFTVLLFDEIEKAHKDINNLMLQL 60
Query 121 LDEGRLTDSQGRTVDFSNCLLILTSNIG 148
LD+G+LTDS G+ VDFSN L+ TSN+G
Sbjct 61 LDDGKLTDSLGKCVDFSNTLIFFTSNLG 88
> ath:AT4G14670 CLPB2; CLPB2; ATP binding / nucleoside-triphosphatase/
nucleotide binding / protein binding
Length=623
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/79 (68%), Positives = 64/79 (81%), Gaps = 0/79 (0%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMS 62
AV AV+ +ILRS GL R +P GSFLFLGPTGVGKTEL KA+AE+LFD++ +VRLDMS
Sbjct 542 AVKAVAAAILRSRVGLGRPQQPSGSFLFLGPTGVGKTELAKALAEQLFDSENLLVRLDMS 601
Query 63 EYMESHSVARLIGAPPGYV 81
EY + SV +LIGAPPGYV
Sbjct 602 EYNDKFSVNKLIGAPPGYV 620
> bbo:BBOV_I001700 19.m02115; chaperone clpB
Length=833
Score = 81.6 bits (200), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 43/125 (34%), Positives = 69/125 (55%), Gaps = 4/125 (3%)
Query 23 KPIGSFLF-LGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
+ IG+ L+ +GP GVGK L V+ +F + + S + S++ L+G+PPGY+
Sbjct 594 RKIGAALYYVGPPGVGKRLLLNHVSR-MFGMALKYIC--GSNLVSSNATNILVGSPPGYI 650
Query 82 GHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDFSNCLL 141
GH EGG L E ++ +PY +V ++ H V N+L+ +D G L D+QG + C
Sbjct 651 GHREGGMLCEWIKDHPYGIVAFEDAHLLHVNVVNLLIGAIDNGFLVDNQGDQCNLRQCFF 710
Query 142 ILTSN 146
+ SN
Sbjct 711 VFVSN 715
> ath:AT4G30350 heat shock protein-related
Length=924
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 64/119 (53%), Gaps = 9/119 (7%)
Query 28 FLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGG 87
+F GP GK+++ A+++ + ++ + L S M+ R G
Sbjct 611 LMFTGPDRAGKSKMASALSDLVSGSQPITISLGSSSRMDDGLNIR---------GKTALD 661
Query 88 QLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDFSNCLLILTSN 146
+ EAVRRNP++V++L+++++A + N + ++ GR+ DS GR V N ++ILT+N
Sbjct 662 RFAEAVRRNPFAVIVLEDIDEADILLRNNVKIAIERGRICDSYGREVSLGNVIIILTAN 720
> ath:AT5G57710 heat shock protein-related
Length=990
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 64/119 (53%), Gaps = 8/119 (6%)
Query 28 FLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGG 87
LF GP VGK ++ A++ ++ T +++L + + + + G
Sbjct 655 LLFSGPDRVGKRKMVSALSSLVYGTNPIMIQLGSRQDAGDGNSS--------FRGKTALD 706
Query 88 QLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDFSNCLLILTSN 146
++ E V+R+P+SV+LL+++++A V + Q +D GR+ DS GR + N + ++T++
Sbjct 707 KIAETVKRSPFSVILLEDIDEADMLVRGSIKQAMDRGRIRDSHGREISLGNVIFVMTAS 765
> ath:AT1G07200 ATP-dependent Clp protease ClpB protein-related
Length=979
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 43/147 (29%), Positives = 67/147 (45%), Gaps = 9/147 (6%)
Query 3 AVTAVSESILRSAAGLARRNKPIGSFL-FLGPTGVGKTELCKAVAEELFDTKERIVRLDM 61
AV A+S+ I RRN+ G +L LGP VGK ++ ++E F K + +D
Sbjct 635 AVNAISQIICGCKTDSTRRNQASGIWLALLGPDKVGKKKVAMTLSEVFFGGKVNYICVDF 694
Query 62 SEYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWNVLLQVL 121
A + G +T + R P+SVVLL+ VEKA L + +
Sbjct 695 G--------AEHCSLDDKFRGKTVVDYVTGELSRKPHSVVLLENVEKAEFPDQMRLSEAV 746
Query 122 DEGRLTDSQGRTVDFSNCLLILTSNIG 148
G++ D GR + N ++++TS I
Sbjct 747 STGKIRDLHGRVISMKNVIVVVTSGIA 773
> dre:792835 torsin family 3, member A
Length=367
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 67/147 (45%), Gaps = 24/147 (16%)
Query 11 ILRSAAGLARR---NKPIGSFLFLGPTGVGKTELCKAVAEELF--DTKERIVRLDMSEYM 65
+L++ G + NKP+ + F G +G GK + + VA+ L+ K VRL ++ +
Sbjct 116 VLKAIQGFIKNPESNKPL-TLSFHGWSGTGKNFVARIVADNLYRDGIKSECVRLFIAPFH 174
Query 66 ESHSVARLIGAPPGYVGHEEGGQLTEAVR----RNPYSVVLLDEVEKAHHQVWNVLLQVL 121
H ARL+ GQL EA+R R P ++ + DE EK H + + + +
Sbjct 175 FPH--ARLVDV--------YKGQLREAIRDMVLRCPQTLFIFDEAEKLHPGLIDAIKPYM 224
Query 122 DEGRLTDSQGRTVDFSNCLLILTSNIG 148
D D V + + + SNIG
Sbjct 225 DHYDNVDG----VSYRRAIFLFLSNIG 247
> dre:100331535 torsin family 3, member A-like
Length=367
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 41/147 (27%), Positives = 67/147 (45%), Gaps = 24/147 (16%)
Query 11 ILRSAAGLARR---NKPIGSFLFLGPTGVGKTELCKAVAEELF--DTKERIVRLDMSEYM 65
+L++ G + NKP+ + F G +G GK + + VA+ L+ K VRL ++ +
Sbjct 116 VLKAIQGFIKNPESNKPL-TLSFHGWSGTGKNFVARIVADNLYRDGIKSECVRLFIAPFH 174
Query 66 ESHSVARLIGAPPGYVGHEEGGQLTEAVR----RNPYSVVLLDEVEKAHHQVWNVLLQVL 121
H ARL+ GQL EA+R R P ++ + DE EK H + + + +
Sbjct 175 FPH--ARLVDV--------YKGQLREAIRDMVLRCPQTLFIFDEAEKLHPGLIDAIKPYM 224
Query 122 DEGRLTDSQGRTVDFSNCLLILTSNIG 148
D D V + + + SNIG
Sbjct 225 DHYDNVDG----VSYRRAIFLFLSNIG 247
> sce:YBL022C PIM1; ATP-dependent Lon protease, involved in degradation
of misfolded proteins in mitochondria; required for
biogenesis and maintenance of mitochondria (EC:3.4.21.-);
K08675 Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=1133
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 40/127 (31%), Positives = 59/127 (46%), Gaps = 16/127 (12%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVAR-LIGAPPGYVGHEEGGQ 88
F+GP GVGKT + K++A L R M++ E R IGA PG V
Sbjct 630 FVGPPGVGKTSIGKSIARALNRKFFRFSVGGMTDVAEIKGHRRTYIGALPGRVVQ----A 685
Query 89 LTEAVRRNPYSVVLLDEVEK-----AHHQVWNVLLQVLDEGR----LTDSQGRTVDFSNC 139
L + +NP ++L+DE++K H LL+VLD + L + +D S
Sbjct 686 LKKCQTQNP--LILIDEIDKIGHGGIHGDPSAALLEVLDPEQNNSFLDNYLDIPIDLSKV 743
Query 140 LLILTSN 146
L + T+N
Sbjct 744 LFVCTAN 750
> dre:100170794 zgc:194342
Length=323
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 55/131 (41%), Gaps = 17/131 (12%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
NKP+ F G G GK + K +A ++ E+ S+++ H+ P
Sbjct 83 NKPL-VLSFHGTAGTGKNHVAKIIARNVYKKGEK------SKHV--HTFISQFHFPHQEN 133
Query 82 GHEEGGQLTEAVRRN----PYSVVLLDEVEKAHHQVWNVLLQVLDEGRLTDSQGRTVDFS 137
H QL + + N P S+ + DE++K H ++ +++ LD D V F
Sbjct 134 VHMYSAQLKQWIHGNVSSFPRSMFIFDEMDKMHPELIDIIKPFLDYNYNVDG----VSFH 189
Query 138 NCLLILTSNIG 148
+ I SN G
Sbjct 190 TAIFIFLSNAG 200
> eco:b0439 lon, capR, deg, dir, ECK0433, JW0429, lonA, lopA,
muc; DNA-binding ATP-dependent protease La (EC:3.4.21.53); K01338
ATP-dependent Lon protease [EC:3.4.21.53]
Length=784
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 45/152 (29%), Positives = 72/152 (47%), Gaps = 20/152 (13%)
Query 7 VSESILRSAAGLARRNKPIGSFLFL-GPTGVGKTELCKAVAEELFDTKERIVRLDMSEYM 65
V + IL A +R NK G L L GP GVGKT L +++A+ T + VR+ +
Sbjct 330 VKDRILEYLAVQSRVNKIKGPILCLVGPPGVGKTSLGQSIAKA---TGRKYVRMALGGVR 386
Query 66 ESHSVARLIGAPPGYVGHEEGGQLTEAVR---RNPYSVVLLDEVEKAHHQV----WNVLL 118
+ A + G Y+G G + + + +NP + LLDE++K + + LL
Sbjct 387 DE---AEIRGHRRTYIGSMPGKLIQKMAKVGVKNP--LFLLDEIDKMSSDMRGDPASALL 441
Query 119 QVLDEGR---LTDSQGRT-VDFSNCLLILTSN 146
+VLD + +D D S+ + + TSN
Sbjct 442 EVLDPEQNVAFSDHYLEVDYDLSDVMFVATSN 473
> dre:563257 lonp1, fc64d11, prss15, wu:fc64d11; lon peptidase
1, mitochondrial; K08675 Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=966
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 47/151 (31%), Positives = 65/151 (43%), Gaps = 16/151 (10%)
Query 7 VSESILRSAAGLARRNKPIGSFL-FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYM 65
V + IL A R G L F GP GVGKT + +++A L R M++
Sbjct 511 VKKRILEFIAVSQLRGSTQGKILCFYGPPGVGKTSIARSIARALNREYFRFSVGGMTDVA 570
Query 66 ESHSVAR-LIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKA----HHQVWNVLLQV 120
E R +GA PG + L + NP +VL+DEV+K + LL++
Sbjct 571 EIKGHRRTYVGAMPGKIIQ----CLKKTKTENP--LVLIDEVDKIGRGYQGDPSSALLEL 624
Query 121 LDEGR----LTDSQGRTVDFSNCLLILTSNI 147
LD + L VD S L I T+NI
Sbjct 625 LDPEQNANFLDHYLDVPVDLSKVLFICTANI 655
> cel:C34B2.6 hypothetical protein; K08675 Lon-like ATP-dependent
protease [EC:3.4.21.-]
Length=971
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 42/129 (32%), Positives = 59/129 (45%), Gaps = 20/129 (15%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGG-- 87
F GP GVGKT + K++A L R M++ VA + G YVG G
Sbjct 510 FHGPPGVGKTSIAKSIATALNREYFRFSVGGMTD------VAEIKGHRRTYVGAMPGKMI 563
Query 88 QLTEAVR-RNPYSVVLLDEVEKA-----HHQVWNVLLQVLDEGRLTDSQGR----TVDFS 137
Q + V+ NP +VL+DEV+K H + LL++LD + + VD S
Sbjct 564 QCMKKVKTENP--LVLIDEVDKIGGAGFHGDPASALLELLDPEQNANFNDHFLDVPVDLS 621
Query 138 NCLLILTSN 146
L I T+N
Sbjct 622 RVLFICTAN 630
> ath:AT2G29970 heat shock protein-related
Length=1002
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 63/149 (42%), Gaps = 18/149 (12%)
Query 3 AVTAVSESILRSAAGLARRNKPIGS-----FLFLGPTGVGKTELCKAVAEELFDTKERIV 57
AV A+SE + RRN + + LGP GK ++ A+AE ++ +
Sbjct 645 AVNAISEIVCGYRDESRRRNNHVATTSNVWLALLGPDKAGKKKVALALAEVFCGGQDNFI 704
Query 58 RLDMS--EYMESHSVARLIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKAHHQVWN 115
+D + ++ + + Y+ E V R SVV ++ VEKA
Sbjct 705 CVDFKSQDSLDDRFRGKTVV---DYIAGE--------VARRADSVVFIENVEKAEFPDQI 753
Query 116 VLLQVLDEGRLTDSQGRTVDFSNCLLILT 144
L + + G+L DS GR + N +++ T
Sbjct 754 RLSEAMRTGKLRDSHGREISMKNVIVVAT 782
> hsa:9361 LONP1, LON, LONP, LonHS, MGC1498, PIM1, PRSS15, hLON;
lon peptidase 1, mitochondrial; K08675 Lon-like ATP-dependent
protease [EC:3.4.21.-]
Length=959
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 45/151 (29%), Positives = 65/151 (43%), Gaps = 16/151 (10%)
Query 7 VSESILRSAAGLARRNKPIGSFL-FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYM 65
V + IL A R G L F GP GVGKT + +++A L R M++
Sbjct 497 VKKRILEFIAVSQLRGSTQGKILCFYGPPGVGKTSIARSIARALNREYFRFSVGGMTDVA 556
Query 66 ESHSVAR-LIGAPPGYVGHEEGGQLTEAVRRNPYSVVLLDEVEKA----HHQVWNVLLQV 120
E R +GA PG + L + NP ++L+DEV+K + LL++
Sbjct 557 EIKGHRRTYVGAMPGKIIQ----CLKKTKTENP--LILIDEVDKIGRGYQGDPSSALLEL 610
Query 121 LDEGR----LTDSQGRTVDFSNCLLILTSNI 147
LD + L VD S L I T+N+
Sbjct 611 LDPEQNANFLDHYLDVPVDLSKVLFICTANV 641
> ath:AT5G47040 LON2; LON2 (LON PROTEASE 2); ATP binding / ATP-dependent
peptidase/ nucleoside-triphosphatase/ nucleotide
binding / serine-type endopeptidase/ serine-type peptidase;
K01338 ATP-dependent Lon protease [EC:3.4.21.53]
Length=888
Score = 37.7 bits (86), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 52/101 (51%), Gaps = 17/101 (16%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGGQL 89
F+GP GVGKT L ++A L + VRL + + A + G Y+G G+L
Sbjct 406 FVGPPGVGKTSLASSIAAAL---GRKFVRLSLGGVKDE---ADIRGHRRTYIGSMP-GRL 458
Query 90 TEAVRR----NPYSVVLLDEVEKAHHQV----WNVLLQVLD 122
+ ++R NP V+LLDE++K V + LL+VLD
Sbjct 459 IDGLKRVGVCNP--VMLLDEIDKTGSDVRGDPASALLEVLD 497
> tgo:TGME49_108580 lon protease, putative (EC:3.4.21.53); K08675
Lon-like ATP-dependent protease [EC:3.4.21.-]
Length=1498
Score = 37.7 bits (86), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 39/126 (30%), Positives = 56/126 (44%), Gaps = 15/126 (11%)
Query 30 FLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVAR-LIGAPPGYVGHEEGGQ 88
+GP GVGKT + +++A L RI M + E R I A PG V
Sbjct 1032 LVGPPGVGKTSVGQSIARALHRKFYRISLGGMCDVAELRGHRRTYISALPGKVIQ----A 1087
Query 89 LTEAVRRNPYSVVLLDEVEKAHHQVW----NVLLQVLDEGRLTDSQGR----TVDFSNCL 140
L E NP V+LLDE++K + LL++LD + + +VD S L
Sbjct 1088 LKECQTMNP--VILLDEIDKLGRDFRGDPSSALLEILDPSQNKSFRDYYLDVSVDLSKVL 1145
Query 141 LILTSN 146
+ T+N
Sbjct 1146 FVCTAN 1151
> bbo:BBOV_III006020 17.m07531; ATP-dependent protease La family
protein
Length=1122
Score = 37.4 bits (85), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 41/127 (32%), Positives = 60/127 (47%), Gaps = 21/127 (16%)
Query 32 GPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVGHEEGGQLTE 91
GP GVGKT + AVAE L ++ R + + VA L G YVG G+ +
Sbjct 638 GPPGVGKTSIATAVAELL---NRKLYRFSLGGLFD---VAELRGHRRTYVG-ALPGKFVQ 690
Query 92 AVR----RNPYSVVLLDEVEK----AHHQVWNVLLQVLDEGRLTDSQGR----TVDFSNC 139
A++ NP +++LDE++K A + LL+VLD + + VD S
Sbjct 691 ALKYTGSMNP--LIVLDEIDKLGRDARGDPASALLEVLDPSQNEYFRDYYLDIPVDLSRV 748
Query 140 LLILTSN 146
L I T+N
Sbjct 749 LFICTAN 755
> tpv:TP04_0031 26S proteasome aaa-ATPase subunit Rpt3 (EC:3.6.4.8);
K03063 26S proteasome regulatory subunit T3
Length=396
Score = 37.4 bits (85), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 32/113 (28%), Positives = 52/113 (46%), Gaps = 25/113 (22%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
+ P+G L+ GP G GKT L KAVA T +R SE+++ + +G P V
Sbjct 174 DPPVGVLLY-GPPGTGKTMLAKAVAHH---TDATFIRFVGSEFVQKY-----LGEGPRMV 224
Query 82 GHEEGGQLTEAVRRNPYSVVLLDEVEK-----------AHHQVWNVLLQVLDE 123
+ R N S++ +DEV+ A +V +LL++L++
Sbjct 225 -----RDIFRLARENAPSILFIDEVDAIATKRFDAQTGADREVQRILLELLNQ 272
> sce:YDR394W RPT3, YNT1, YTA2; One of six ATPases of the 19S
regulatory particle of the 26S proteasome involved in the degradation
of ubiquitinated substrates; substrate of N-acetyltransferase
B; K03063 26S proteasome regulatory subunit T3
Length=428
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 44/90 (48%), Gaps = 22/90 (24%)
Query 22 NKPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYV 81
+ P G L+ GP G GKT L KAVA TK +R++ SE++ Y+
Sbjct 204 DPPRGVLLY-GPPGTGKTMLVKAVANS---TKAAFIRVNGSEFVHK------------YL 247
Query 82 GHEEGGQLTEAV----RRNPYSVVLLDEVE 107
G EG ++ V R N S++ +DEV+
Sbjct 248 G--EGPRMVRDVFRLARENAPSIIFIDEVD 275
> ath:AT1G62130 AAA-type ATPase family protein
Length=1025
Score = 37.4 bits (85), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 41/95 (43%), Gaps = 27/95 (28%)
Query 23 KPIGSFLFLGPTGVGKTELCKAVAEELFDTKERIVRLDMSEYMESHSVARLIGAPPGYVG 82
KP L GP+G GKT L KAVA E ++ + MS +
Sbjct 768 KPCNGILLFGPSGTGKTMLAKAVATE---AGANLINMSMSRWF----------------- 807
Query 83 HEEGGQLTEAV-----RRNPYSVVLLDEVEKAHHQ 112
EG + +AV + +P S++ LDEVE H+
Sbjct 808 -SEGEKYVKAVFSLASKISP-SIIFLDEVESMLHR 840
Lambda K H
0.318 0.135 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3003468616
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40