bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5446_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
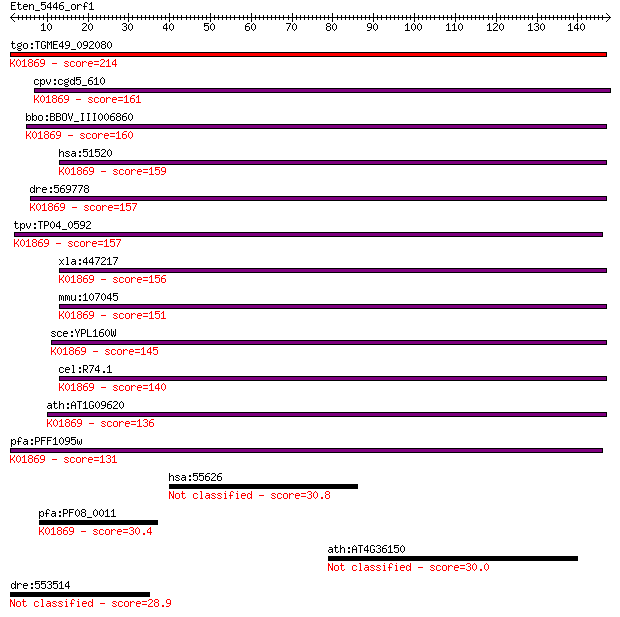
tgo:TGME49_092080 leucyl-tRNA synthetase, putative (EC:6.1.1.4... 214 1e-55
cpv:cgd5_610 hypothetical protein ; K01869 leucyl-tRNA synthet... 161 8e-40
bbo:BBOV_III006860 17.m07606; leucyl-tRNA synthetase; K01869 l... 160 2e-39
hsa:51520 LARS, FLJ10595, FLJ21788, HSPC192, KIAA1352, LARS1, ... 159 3e-39
dre:569778 lars, cb667, wu:fb36b12; leucyl-tRNA synthetase; K0... 157 1e-38
tpv:TP04_0592 leucyl-tRNA synthetase (EC:6.1.1.4); K01869 leuc... 157 2e-38
xla:447217 lars, MGC82093; leucyl-tRNA synthetase (EC:6.1.1.4)... 156 3e-38
mmu:107045 Lars, 2310045K21Rik, 3110009L02Rik, AW536573, mKIAA... 151 7e-37
sce:YPL160W CDC60; Cdc60p (EC:6.1.1.4); K01869 leucyl-tRNA syn... 145 4e-35
cel:R74.1 lrs-1; Leucyl tRNA Synthetase family member (lrs-1);... 140 1e-33
ath:AT1G09620 ATP binding / aminoacyl-tRNA ligase/ leucine-tRN... 136 2e-32
pfa:PFF1095w leucyl tRNA synthase (EC:6.1.1.4); K01869 leucyl-... 131 8e-31
hsa:55626 AMBRA1, DCAF3, FLJ20294, KIAA1736, MGC33725, WDR94; ... 30.8 1.7
pfa:PF08_0011 leucine-tRNA ligase; K01869 leucyl-tRNA syntheta... 30.4 2.3
ath:AT4G36150 disease resistance protein (TIR-NBS-LRR class), ... 30.0 2.8
dre:553514 im:7138204 28.9 5.5
> tgo:TGME49_092080 leucyl-tRNA synthetase, putative (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=1162
Score = 214 bits (544), Expect = 1e-55, Method: Composition-based stats.
Identities = 92/146 (63%), Positives = 112/146 (76%), Gaps = 0/146 (0%)
Query 1 HLTSGHFDPHSPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTI 60
++ S F SPQ +HQF V+ WL+EWACSRSYGLGT+LPWT D+ PVLIESLSDSTI
Sbjct 616 YINSEQFQTFSPQVLHQFKHVVGWLREWACSRSYGLGTYLPWTKDSSRPVLIESLSDSTI 675
Query 61 YMAYYTVAHLMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRL 120
YMAYY +AHL+QG D+YG GPLG+ LT++ FDY+F D+ PK + + E+L R+
Sbjct 676 YMAYYAIAHLLQGNDMYGQAKGPLGIAVEQLTDEVFDYVFAQTDDLPKGSTIPAEHLKRM 735
Query 121 RAEFSYWYPMDLRVSGKDLIFNHLTF 146
R EF YWYP+DLRVSGKDLIFNHLTF
Sbjct 736 RNEFEYWYPLDLRVSGKDLIFNHLTF 761
> cpv:cgd5_610 hypothetical protein ; K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=632
Score = 161 bits (407), Expect = 8e-40, Method: Composition-based stats.
Identities = 76/143 (53%), Positives = 97/143 (67%), Gaps = 3/143 (2%)
Query 7 FDPHSPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYT 66
F+ PQ F V++WL+EWACSRSYGLGT+LPW T+ + VLIESLSDSTIYMAYYT
Sbjct 295 FETFYPQVRTSFLEVINWLREWACSRSYGLGTYLPWDTENNQKVLIESLSDSTIYMAYYT 354
Query 67 VAHLMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKD--TKVSLENLNRLRAEF 124
+ H D G + G + + + + F+Y+F L DEP +D + L+R+R EF
Sbjct 355 ICHFFH-SDFEGRSKGLMDIPIEYVNDDLFNYVFCLTDEPSEDLIKNIGRGQLDRMRNEF 413
Query 125 SYWYPMDLRVSGKDLIFNHLTFC 147
SY+YP+D RVSGKDLIFNHLT C
Sbjct 414 SYFYPLDCRVSGKDLIFNHLTMC 436
> bbo:BBOV_III006860 17.m07606; leucyl-tRNA synthetase; K01869
leucyl-tRNA synthetase [EC:6.1.1.4]
Length=1098
Score = 160 bits (405), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 79/144 (54%), Positives = 98/144 (68%), Gaps = 3/144 (2%)
Query 5 GHFDPHSPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWT-TDTDAPVLIESLSDSTIYMA 63
F ++ ++Q V+SWL WACSRSYGLGT LPW + + LIESLSDSTIYMA
Sbjct 582 NKFSCYNDSALNQVKHVVSWLDNWACSRSYGLGTVLPWEDINQNKNSLIESLSDSTIYMA 641
Query 64 YYTVAHLMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKD-TKVSLENLNRLRA 122
YYT+AH +QG DI+G TPG LGL P+ LT FDY+F + D P + + L +R
Sbjct 642 YYTIAHYLQG-DIFGTTPGSLGLVPSQLTPAVFDYVFHISDTVPDGLDNATAKKLKLMRD 700
Query 123 EFSYWYPMDLRVSGKDLIFNHLTF 146
+F+YWYP+DLR SGKDLIFNHLT
Sbjct 701 DFAYWYPVDLRCSGKDLIFNHLTM 724
> hsa:51520 LARS, FLJ10595, FLJ21788, HSPC192, KIAA1352, LARS1,
LEURS, LEUS, LRS, PIG44, RNTLS, hr025Cl; leucyl-tRNA synthetase
(EC:6.1.1.4); K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=1176
Score = 159 bits (402), Expect = 3e-39, Method: Composition-based stats.
Identities = 72/134 (53%), Positives = 99/134 (73%), Gaps = 5/134 (3%)
Query 13 QTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHLMQ 72
+T F L WL+E ACSR+YGLGT LPW D LIESLSDSTIYMA+YTVAHL+Q
Sbjct 556 ETRRNFEATLGWLQEHACSRTYGLGTHLPW----DEQWLIESLSDSTIYMAFYTVAHLLQ 611
Query 73 GGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDL 132
GG+++G PLG+ P +T++ +DY+F ++ P T+++ E L++L+ EF +WYP+DL
Sbjct 612 GGNLHGQAESPLGIRPQQMTKEVWDYVF-FKEAPFPKTQIAKEKLDQLKQEFEFWYPVDL 670
Query 133 RVSGKDLIFNHLTF 146
RVSGKDL+ NHL++
Sbjct 671 RVSGKDLVPNHLSY 684
> dre:569778 lars, cb667, wu:fb36b12; leucyl-tRNA synthetase;
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=747
Score = 157 bits (398), Expect = 1e-38, Method: Composition-based stats.
Identities = 76/141 (53%), Positives = 96/141 (68%), Gaps = 5/141 (3%)
Query 6 HFDPHSPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYY 65
H + +T F L+WL+E ACSR+YGLGT LPW LIESLSDSTIYMAYY
Sbjct 121 HLETFCDETRKNFEATLAWLQEHACSRTYGLGTRLPWNEQW----LIESLSDSTIYMAYY 176
Query 66 TVAHLMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFS 125
TVAHL+QGG + G P PLG+ P +T + +DYIF PK T + E+L +LR EF
Sbjct 177 TVAHLLQGGVLSGQGPSPLGIRPQQMTREVWDYIFFKTSPFPK-TDIPKEHLLKLRREFE 235
Query 126 YWYPMDLRVSGKDLIFNHLTF 146
+WYP+D+RVSGKDL+ NHL++
Sbjct 236 FWYPVDVRVSGKDLVPNHLSY 256
> tpv:TP04_0592 leucyl-tRNA synthetase (EC:6.1.1.4); K01869 leucyl-tRNA
synthetase [EC:6.1.1.4]
Length=1158
Score = 157 bits (396), Expect = 2e-38, Method: Composition-based stats.
Identities = 79/156 (50%), Positives = 100/156 (64%), Gaps = 13/156 (8%)
Query 2 LTSGHFDPHSPQTMHQFSFVLSWLKEWACSRSYGLGTFLPW-TTDTDAPVLIESLSDSTI 60
+TS F ++ T +Q ++ WL WACSRSYGLGT LPW + +LIESLSDSTI
Sbjct 575 VTSNQFTCYNESTYNQLLNIIQWLDNWACSRSYGLGTLLPWENIKNNKNILIESLSDSTI 634
Query 61 YMAYYTVAHLMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKV---SLEN- 116
YMAYYTVAH +Q DI+G PG L L+ + FDYIF + D P T +LEN
Sbjct 635 YMAYYTVAHYLQS-DIFGTNPGLLNLSSKQINYSLFDYIFRISDNLPSLTHTGQGNLENH 693
Query 117 -------LNRLRAEFSYWYPMDLRVSGKDLIFNHLT 145
+NR+R EF+YWYP+++R SGKDL+FNHLT
Sbjct 694 RDDVMDKINRMREEFNYWYPVNVRCSGKDLLFNHLT 729
> xla:447217 lars, MGC82093; leucyl-tRNA synthetase (EC:6.1.1.4);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=1178
Score = 156 bits (394), Expect = 3e-38, Method: Composition-based stats.
Identities = 74/134 (55%), Positives = 94/134 (70%), Gaps = 5/134 (3%)
Query 13 QTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHLMQ 72
+T F L WL+E ACSR+YGLG+ LPW D LIESLSDSTIYMAYYTV HL+Q
Sbjct 558 ETRRNFEATLGWLQEHACSRTYGLGSRLPW----DEQWLIESLSDSTIYMAYYTVCHLLQ 613
Query 73 GGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDL 132
G D+ G PLG+ P +T++ +DYIF + PK T + E LN+L+ EF +WYP+DL
Sbjct 614 GKDLSGQGESPLGIRPEQMTKEVWDYIFFKKAPFPK-TTIQKEKLNKLKQEFEFWYPVDL 672
Query 133 RVSGKDLIFNHLTF 146
RVSGKDL+ NHL++
Sbjct 673 RVSGKDLVPNHLSY 686
> mmu:107045 Lars, 2310045K21Rik, 3110009L02Rik, AW536573, mKIAA1352;
leucyl-tRNA synthetase (EC:6.1.1.4); K01869 leucyl-tRNA
synthetase [EC:6.1.1.4]
Length=1178
Score = 151 bits (382), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 73/134 (54%), Positives = 95/134 (70%), Gaps = 5/134 (3%)
Query 13 QTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHLMQ 72
++ F L WL+E ACSR+YGLGT LPW D LIESLSDSTIYMA+YTVAHL+Q
Sbjct 558 ESRKNFEASLDWLQEHACSRTYGLGTRLPW----DEQWLIESLSDSTIYMAFYTVAHLLQ 613
Query 73 GGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDL 132
GGD+ G PLG+ P +T+ +DY+F +D P T++ E L++L+ EF +WYP+DL
Sbjct 614 GGDLNGQAESPLGIRPQQMTKDVWDYVF-FKDAPFPKTQIPKEKLDQLKQEFEFWYPVDL 672
Query 133 RVSGKDLIFNHLTF 146
R SGKDLI NHL++
Sbjct 673 RASGKDLIPNHLSY 686
> sce:YPL160W CDC60; Cdc60p (EC:6.1.1.4); K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=1090
Score = 145 bits (366), Expect = 4e-35, Method: Composition-based stats.
Identities = 71/136 (52%), Positives = 95/136 (69%), Gaps = 6/136 (4%)
Query 11 SPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHL 70
+P+ + F VL WLK WA R+YGLGT LPW D L+ESLSDSTIY ++YT+AHL
Sbjct 570 APEVKNAFEGVLDWLKNWAVCRTYGLGTRLPW----DEKYLVESLSDSTIYQSFYTIAHL 625
Query 71 MQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPM 130
+ D YG+ GPLG++ +T++ FDYIF +D+ K+T + L L +LR EF Y+YP+
Sbjct 626 L-FKDYYGNEIGPLGISADQMTDEVFDYIFQHQDDV-KNTNIPLPALQKLRREFEYFYPL 683
Query 131 DLRVSGKDLIFNHLTF 146
D+ +SGKDLI NHLTF
Sbjct 684 DVSISGKDLIPNHLTF 699
> cel:R74.1 lrs-1; Leucyl tRNA Synthetase family member (lrs-1);
K01869 leucyl-tRNA synthetase [EC:6.1.1.4]
Length=1186
Score = 140 bits (354), Expect = 1e-33, Method: Composition-based stats.
Identities = 71/134 (52%), Positives = 83/134 (61%), Gaps = 4/134 (2%)
Query 13 QTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAHLMQ 72
+T + WL E+ACSRSYGLGT LPW D LIESLSDSTIY AYYTVAHL+Q
Sbjct 557 ETRRGLETTVDWLHEYACSRSYGLGTKLPW----DTQYLIESLSDSTIYNAYYTVAHLLQ 612
Query 73 GGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDL 132
G G GP G+ +T+ + Y+FL K V E L LR EF YWYP+D+
Sbjct 613 QGAFDGSVVGPAGIKADQMTDASWSYVFLGEIYDSKTMPVEEEKLKSLRKEFMYWYPIDM 672
Query 133 RVSGKDLIFNHLTF 146
R SGKDLI NHLT+
Sbjct 673 RASGKDLIGNHLTY 686
> ath:AT1G09620 ATP binding / aminoacyl-tRNA ligase/ leucine-tRNA
ligase/ nucleotide binding (EC:6.1.1.4); K01869 leucyl-tRNA
synthetase [EC:6.1.1.4]
Length=1091
Score = 136 bits (343), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 69/138 (50%), Positives = 93/138 (67%), Gaps = 9/138 (6%)
Query 10 HSPQTMHQFSFVLSWLKEWACSRSYGLGTFLPWTTDTDAPVLIESLSDSTIYMAYYTVAH 69
+S +T H F LSWL +WACSRS+GLGT +PW D L+ESLSDS++YMAYYTVAH
Sbjct 556 YSDETRHGFEHTLSWLNQWACSRSFGLGTRIPW----DEQFLVESLSDSSLYMAYYTVAH 611
Query 70 LMQGGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEP-PKDTKVSLENLNRLRAEFSYWY 128
+ GD+Y + + P + ++ ++Y+F D P PK + + L+ ++ EF YWY
Sbjct 612 IFHDGDMYKGSKSLI--RPQQMNDEVWEYLFC--DGPYPKSSDIPSAVLSEMKQEFDYWY 667
Query 129 PMDLRVSGKDLIFNHLTF 146
P+DLRVSGKDLI NHLTF
Sbjct 668 PLDLRVSGKDLIQNHLTF 685
> pfa:PFF1095w leucyl tRNA synthase (EC:6.1.1.4); K01869 leucyl-tRNA
synthetase [EC:6.1.1.4]
Length=1447
Score = 131 bits (329), Expect = 8e-31, Method: Composition-based stats.
Identities = 75/194 (38%), Positives = 98/194 (50%), Gaps = 50/194 (25%)
Query 1 HLTSGHFDPHSPQTMHQFSFVLSWLKEWACSRSYGLGTF--------------------- 39
L +F ++ Q V+ WL +W+CSRSYGLGT+
Sbjct 724 QLKKNNFQTYNDVLYKQLQHVIFWLDDWSCSRSYGLGTYMPQFDQNNQTNKNVDNYQNNN 783
Query 40 ---LPWTTDTDAPV------------------------LIESLSDSTIYMAYYTVAHLMQ 72
+P D + LIESLSDSTIYMAYYTV+H +Q
Sbjct 784 EYVIPSNDDNNQINNHHINVVKEEEEKNVHIKNNSRKELIESLSDSTIYMAYYTVSHFLQ 843
Query 73 GGDIYGDTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTK-VSLENLNRLRAEFSYWYPMD 131
G + G G L ++ DL + FFDYIF + D+ +K +S E L R+R EF+YWYP D
Sbjct 844 G-SVDGQKRGLLDISADDLNDAFFDYIFDISDDTSNISKHISKEKLVRMRREFTYWYPFD 902
Query 132 LRVSGKDLIFNHLT 145
+R+SGKDLIFNHLT
Sbjct 903 VRISGKDLIFNHLT 916
> hsa:55626 AMBRA1, DCAF3, FLJ20294, KIAA1736, MGC33725, WDR94;
autophagy/beclin-1 regulator 1
Length=1208
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 15/46 (32%), Positives = 21/46 (45%), Gaps = 0/46 (0%)
Query 40 LPWTTDTDAPVLIESLSDSTIYMAYYTVAHLMQGGDIYGDTPGPLG 85
LP ++ PV + S T++ HL+ GG GD GP G
Sbjct 1158 LPSSSPVPIPVSLPSAEGPTLHCELTNNNHLLDGGSSRGDAAGPRG 1203
> pfa:PF08_0011 leucine-tRNA ligase; K01869 leucyl-tRNA synthetase
[EC:6.1.1.4]
Length=1481
Score = 30.4 bits (67), Expect = 2.3, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 8 DPHSPQTMHQFSFVLSWLKEWACSRSYGL 36
DPH+ Q ++ S VLS K+W ++ Y +
Sbjct 799 DPHNEQDVNNISSVLSRFKKWMITKKYNM 827
> ath:AT4G36150 disease resistance protein (TIR-NBS-LRR class),
putative
Length=1179
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 3/61 (4%)
Query 79 DTPGPLGLTPADLTEQFFDYIFLLRDEPPKDTKVSLENLNRLRAEFSYWYPMDLRVSGKD 138
DT L LT D E F + F + PP T NL+RL A+++ P+ L++ GK+
Sbjct 363 DTYEVLRLTGRDSFEYFSYFAFSGKLCPPVRT---FMNLSRLFADYAKGNPLALKILGKE 419
Query 139 L 139
L
Sbjct 420 L 420
> dre:553514 im:7138204
Length=1652
Score = 28.9 bits (63), Expect = 5.5, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 19/34 (55%), Gaps = 0/34 (0%)
Query 1 HLTSGHFDPHSPQTMHQFSFVLSWLKEWACSRSY 34
H+ + F+ H P T+ F + L E+AC+ S+
Sbjct 1111 HIVTNVFEKHFPATIDSFQDAVKCLSEFACNASF 1144
Lambda K H
0.322 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40