bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5436_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
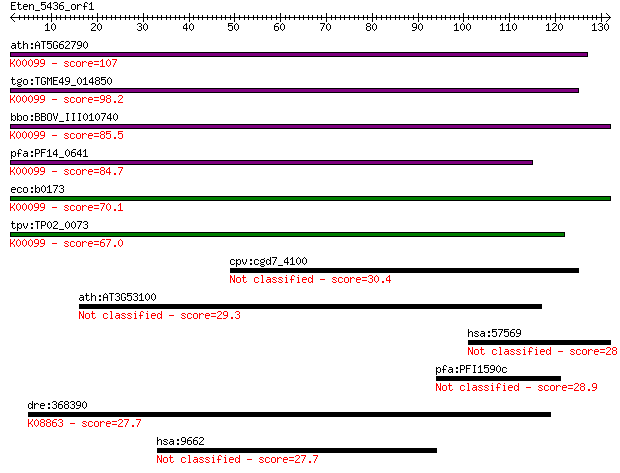
ath:AT5G62790 DXR; DXR (1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTO... 107 7e-24
tgo:TGME49_014850 1-deoxy-D-xylulose 5-phosphate reductoisomer... 98.2 5e-21
bbo:BBOV_III010740 17.m07925; 1-deoxy-D-xylulose 5-phosphate r... 85.5 4e-17
pfa:PF14_0641 1-deoxy-D-xylulose 5-phosphate reductoisomerase;... 84.7 7e-17
eco:b0173 dxr, ECK0172, ispC, JW0168, yaeM; 1-deoxy-D-xylulose... 70.1 2e-12
tpv:TP02_0073 1-deoxy-D-xylulose 5-phosphate reductoisomerase;... 67.0 1e-11
cpv:cgd7_4100 Ssm4 ring finger fused to a forkhead associated ... 30.4 1.4
ath:AT3G53100 GDSL-motif lipase/hydrolase family protein 29.3 3.3
hsa:57569 ARHGAP20, KIAA1391, RARHOGAP; Rho GTPase activating ... 28.9 3.9
pfa:PFI1590c conserved Plasmodium protein, unknown function 28.9 4.2
dre:368390 plk4, stk18, wu:fi33g07; polo-like kinase 4 (Drosop... 27.7 9.1
hsa:9662 CEP135, CEP4, KIAA0635; centrosomal protein 135kDa 27.7 9.1
> ath:AT5G62790 DXR; DXR (1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE);
1-deoxy-D-xylulose-5-phosphate reductoisomerase
(EC:1.1.1.267); K00099 1-deoxy-D-xylulose-5-phosphate reductoisomerase
[EC:1.1.1.267]
Length=477
Score = 107 bits (268), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 57/130 (43%), Positives = 83/130 (63%), Gaps = 4/130 (3%)
Query 1 DRSVMAQLGTADMRLPIQYALTYPQRL--ARETAERLNLWELGKRHFGKLDMERYRCMKF 58
D SV+AQLG DMRLPI Y +++P R+ + T RL+L +LG F K D +Y M
Sbjct 333 DSSVLAQLGWPDMRLPILYTMSWPDRVPCSEVTWPRLDLCKLGSLTFKKPDNVKYPSMDL 392
Query 59 AYDAGRAAGSMPTVLNAANEVAVAAFLNNQISFLQIEDLIEASLNQHVR--ISEPSLDEI 116
AY AGRA G+M VL+AANE AV F++ +IS+L I ++E + ++H ++ PSL+EI
Sbjct 393 AYAAGRAGGTMTGVLSAANEKAVEMFIDEKISYLDIFKVVELTCDKHRNELVTSPSLEEI 452
Query 117 LEADAKTRQF 126
+ D R++
Sbjct 453 VHYDLWAREY 462
> tgo:TGME49_014850 1-deoxy-D-xylulose 5-phosphate reductoisomerase,
putative (EC:1.1.1.267); K00099 1-deoxy-D-xylulose-5-phosphate
reductoisomerase [EC:1.1.1.267]
Length=513
Score = 98.2 bits (243), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 53/128 (41%), Positives = 72/128 (56%), Gaps = 4/128 (3%)
Query 1 DRSVMAQLGTADMRLPIQYALTYPQRLARETAERLNLWELGKRHFGKLDMERYRCMKFAY 60
D + +AQLG DM+LPI YALT+P RLA + ++L G F K D+ + C+ AY
Sbjct 376 DGATLAQLGLPDMKLPIAYALTWPHRLAAPWSAGVDLTREGNLTFEKPDLNTFGCLGLAY 435
Query 61 DAGRAAGSMPTVLNAANEVAVAAFLNNQISFLQIEDLIEASLNQHVR----ISEPSLDEI 116
+AG G P LNAANEVAV F N +I F+ IED + + R S+ SL ++
Sbjct 436 EAGERGGVAPACLNAANEVAVERFRNKEIGFVDIEDTVRHVMALQERERDNFSDVSLQDV 495
Query 117 LEADAKTR 124
+AD R
Sbjct 496 FDADHWAR 503
> bbo:BBOV_III010740 17.m07925; 1-deoxy-D-xylulose 5-phosphate
reductoisomerase family protein (EC:1.1.1.267); K00099 1-deoxy-D-xylulose-5-phosphate
reductoisomerase [EC:1.1.1.267]
Length=447
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 47/133 (35%), Positives = 78/133 (58%), Gaps = 4/133 (3%)
Query 1 DRSVMAQLGTADMRLPIQYALTYPQRLARETAERLNLWELGKRHFGKLDMERYRCMKFAY 60
D SV+AQ+ DMRLPI YAL +P R+ T L+L + + F D+E++ C+K A+
Sbjct 313 DNSVLAQMYNPDMRLPIAYALNWPDRM-ESTLPELDLLK-QQLTFTDPDLEKFPCLKLAF 370
Query 61 DAGRAAGSMPTVLNAANEVAVAAFLNNQISFLQIEDLIEASLNQ--HVRISEPSLDEILE 118
+AG+ G TVLNAANE A + I+ +I +L++ ++ + H I P +++++E
Sbjct 371 EAGKMGGLYTTVLNAANERANEFLREDAIAHWEIHELVKRAVEEYKHPNIDHPRIEDVME 430
Query 119 ADAKTRQFVQSKL 131
AD+ + V +
Sbjct 431 ADSWGKNHVMESM 443
> pfa:PF14_0641 1-deoxy-D-xylulose 5-phosphate reductoisomerase;
K00099 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC:1.1.1.267]
Length=488
Score = 84.7 bits (208), Expect = 7e-17, Method: Composition-based stats.
Identities = 45/116 (38%), Positives = 69/116 (59%), Gaps = 3/116 (2%)
Query 1 DRSVMAQLGTADMRLPIQYALTYPQRLARETAERLNLWELGKRHFGKLDMERYRCMKFAY 60
D+SV++Q+ DM++PI Y+LT+P R+ + + L+L ++ F K +E + C+K AY
Sbjct 348 DKSVISQMYYPDMQIPILYSLTWPDRI-KTNLKPLDLAQVSTLTFHKPSLEHFPCIKLAY 406
Query 61 DAGRAAGSMPTVLNAANEVAVAAFLNNQISFLQIEDLIEASLN--QHVRISEPSLD 114
AG PTVLNA+NE+A FLNN+I + I +I L ++SE S D
Sbjct 407 QAGIKGNFYPTVLNASNEIANNLFLNNKIKYFDISSIISQVLESFNSQKVSENSED 462
> eco:b0173 dxr, ECK0172, ispC, JW0168, yaeM; 1-deoxy-D-xylulose
5-phosphate reductoisomerase (EC:1.1.1.267); K00099 1-deoxy-D-xylulose-5-phosphate
reductoisomerase [EC:1.1.1.267]
Length=398
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 49/132 (37%), Positives = 73/132 (55%), Gaps = 3/132 (2%)
Query 1 DRSVMAQLGTADMRLPIQYALTYPQRLARETAERLNLWELGKRHFGKLDMERYRCMKFAY 60
D SV+AQLG DMR PI + + +P R+ + L+ +L F D +RY C+K A
Sbjct 264 DGSVLAQLGEPDMRTPIAHTMAWPNRV-NSGVKPLDFCKLSALTFAAPDYDRYPCLKLAM 322
Query 61 DAGRAAGSMPTVLNAANEVAVAAFLNNQISFLQIEDLIEASLNQHVRISEP-SLDEILEA 119
+A + T LNAANE+ VAAFL QI F I L S+ + + + EP +D++L
Sbjct 323 EAFEQGQAATTALNAANEITVAAFLAQQIRFTDIAAL-NLSVLEKMDMREPQCVDDVLSV 381
Query 120 DAKTRQFVQSKL 131
DA R+ + ++
Sbjct 382 DANAREVARKEV 393
> tpv:TP02_0073 1-deoxy-D-xylulose 5-phosphate reductoisomerase;
K00099 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC:1.1.1.267]
Length=401
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 41/125 (32%), Positives = 67/125 (53%), Gaps = 8/125 (6%)
Query 1 DRSVMAQLGTADMRLPIQYALTYPQRLARETAERLNLWELGKRH--FGKLDMERYRCMKF 58
D SV+ Q+ DM+LPI +AL +P R T L +L R+ F + D E + +
Sbjct 266 DNSVLCQMYLPDMKLPISHALHWPNR----TKNALPSLDLRGRNLSFKEADYENFPFLGL 321
Query 59 AYDAGRAAGSMPTVLNAANEVAVAAFLNNQISFLQIEDLIEASLNQHVR--ISEPSLDEI 116
Y+ GR G P V NAAN+VA F +N+I + ++ ++ ++ + +S+ L +I
Sbjct 322 GYEVGRMGGLYPAVFNAANDVANGLFRSNKIDYEKLYQIVRETVESFNKPDLSDDILQDI 381
Query 117 LEADA 121
L AD+
Sbjct 382 LYADS 386
> cpv:cgd7_4100 Ssm4 ring finger fused to a forkhead associated
(FHA) domain (apicomplexan specific architecture)
Length=726
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 35/78 (44%), Gaps = 2/78 (2%)
Query 49 DMERYRCMKFAYDAGRAAGSMPTVLNAANEVAVAAFLNNQIS-FLQIEDLIEASLNQHVR 107
D + +K D + S PT NA E + LNNQ + L+IED LN V+
Sbjct 274 DFQDNSLVKCENDISKFMASSPTSNNATREDLGSQILNNQDNQALKIEDCDPTVLNPSVK 333
Query 108 IS-EPSLDEILEADAKTR 124
+ D ++ D K++
Sbjct 334 AQIQTKFDRVINCDQKSK 351
> ath:AT3G53100 GDSL-motif lipase/hydrolase family protein
Length=351
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 47/114 (41%), Gaps = 23/114 (20%)
Query 16 PIQYALTYPQRLA----RETAERL-NLWELGKRHFGKLDMERYRCMKFAYDAGRAAGSMP 70
P+ L P + A R +E + NL+ELG R G + + C+ A
Sbjct 178 PLLNILNTPDQFADILLRSFSEFIQNLYELGARRIGVISLPPMGCLPAAI---------- 227
Query 71 TVLNAANEVAVAAFLNNQISF-LQIEDLIEASLNQH-------VRISEPSLDEI 116
T+ A N+ V N+ I F ++E+ +N+H + +P LD I
Sbjct 228 TLFGAGNKSCVERLNNDAIMFNTKLENTTRLLMNRHSGLRLVAFNVYQPFLDII 281
> hsa:57569 ARHGAP20, KIAA1391, RARHOGAP; Rho GTPase activating
protein 20
Length=1191
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 101 SLNQHVRISEPSLDEILEADAKTRQFVQSKL 131
SL +H R SEPS+D + + R+F Q KL
Sbjct 696 SLRRHRRCSEPSIDYLDSKLSYLREFYQKKL 726
> pfa:PFI1590c conserved Plasmodium protein, unknown function
Length=1342
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 19/27 (70%), Gaps = 1/27 (3%)
Query 94 IEDLIEASLNQHVRISEPSLDEILEAD 120
+E +IE LN H+R+ +DEI+EAD
Sbjct 657 VEKVIERVLN-HIRLKNIDIDEIIEAD 682
> dre:368390 plk4, stk18, wu:fi33g07; polo-like kinase 4 (Drosophila)
(EC:2.7.11.21); K08863 polo-like kinase 4 [EC:2.7.11.21]
Length=941
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 30/124 (24%), Positives = 51/124 (41%), Gaps = 24/124 (19%)
Query 5 MAQLGTA-DMRLPIQYALTY---PQRLARETAERL------NLWELGKRHFGKLDMERYR 54
+A G A ++LP + T P ++ E A R ++W LG
Sbjct 152 IADFGLATQLKLPSEKHFTMCGTPNYISPEVATRSAHGLESDVWSLG------------- 198
Query 55 CMKFAYDAGRAAGSMPTVLNAANEVAVAAFLNNQISFLQIEDLIEASLNQHVRISEPSLD 114
CM +A+ GR TV N+V + + + +DLI+ L ++ + PSL
Sbjct 199 CMFYAFLTGRPPFDTDTVKRTLNKVVLGEYQMPMHISAEAQDLIQQLLQKNPAL-RPSLS 257
Query 115 EILE 118
+L+
Sbjct 258 AVLD 261
> hsa:9662 CEP135, CEP4, KIAA0635; centrosomal protein 135kDa
Length=1140
Score = 27.7 bits (60), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 34/72 (47%), Gaps = 19/72 (26%)
Query 33 ERLNLWELGKRHFGKL------DMERYRCMKFAYDAGRAAGSMPTVL-----NAANEVAV 81
E+L + E G R + K ++ER + A D GR+ P VL N NE +
Sbjct 218 EKLAMMESGVRDYSKQIELREREIER---LSVALDGGRS----PDVLSLESRNKTNEKLI 270
Query 82 AAFLNNQISFLQ 93
A LN Q+ FLQ
Sbjct 271 A-HLNIQVDFLQ 281
Lambda K H
0.320 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40