bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5409_orf1
Length=69
Score E
Sequences producing significant alignments: (Bits) Value
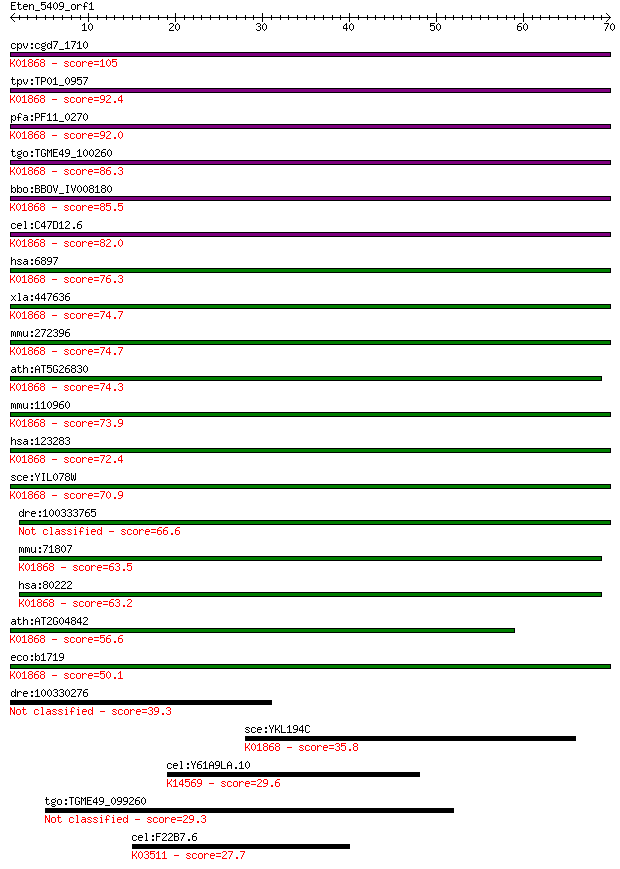
cpv:cgd7_1710 threonyl-tRNA synthetase (RNA binding domain TGS... 105 5e-23
tpv:TP01_0957 threonyl-tRNA synthetase; K01868 threonyl-tRNA s... 92.4 3e-19
pfa:PF11_0270 threonine --tRNA ligase, putative; K01868 threon... 92.0 4e-19
tgo:TGME49_100260 threonyl-tRNA synthetase, putative (EC:6.1.1... 86.3 2e-17
bbo:BBOV_IV008180 23.m05860; threonyl-tRNA synthetase (EC:6.1.... 85.5 3e-17
cel:C47D12.6 trs-1; Threonyl tRNA Synthetase family member (tr... 82.0 4e-16
hsa:6897 TARS, MGC9344, ThrRS; threonyl-tRNA synthetase (EC:6.... 76.3 2e-14
xla:447636 tars, MGC86352; threonyl-tRNA synthetase (EC:6.1.1.... 74.7 7e-14
mmu:272396 Tarsl2, A530046H20Rik, MGC31414; threonyl-tRNA synt... 74.7 7e-14
ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligas... 74.3 9e-14
mmu:110960 Tars, D15Wsu59e, ThrRS; threonyl-tRNA synthetase (E... 73.9 1e-13
hsa:123283 TARSL2, FLJ25005; threonyl-tRNA synthetase-like 2 (... 72.4 3e-13
sce:YIL078W THS1; Ths1p (EC:6.1.1.3); K01868 threonyl-tRNA syn... 70.9 9e-13
dre:100333765 Threonyl-tRNA synthetase, cytoplasmic-like 66.6 2e-11
mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRN... 63.5 2e-10
hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase 2,... 63.2 2e-10
ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding / a... 56.6 2e-08
eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:... 50.1 2e-06
dre:100330276 threonyl-tRNA synthetase-like 39.3 0.003
sce:YKL194C MST1; Mitochondrial threonyl-tRNA synthetase (EC:6... 35.8 0.039
cel:Y61A9LA.10 hypothetical protein; K14569 ribosome biogenesi... 29.6 2.6
tgo:TGME49_099260 hypothetical protein 29.3 3.2
cel:F22B7.6 polk-1; POLK (DNA polymerase kappa) homolog family... 27.7 9.6
> cpv:cgd7_1710 threonyl-tRNA synthetase (RNA binding domain TGS+HxxxH+tRNA
synthetase) ; K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=763
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 46/69 (66%), Positives = 59/69 (85%), Gaps = 0/69 (0%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YGVSFP+KK+L EYL++LE+AK+RDHRLLG+NL LFFF+ VS G FWLP GA++YN L
Sbjct 310 YGVSFPDKKRLDEYLNMLEEAKKRDHRLLGSNLQLFFFDSNVSPGSCFWLPAGARLYNKL 369
Query 61 IEFIREEYQ 69
++FIR EY+
Sbjct 370 MDFIRNEYR 378
> tpv:TP01_0957 threonyl-tRNA synthetase; K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=779
Score = 92.4 bits (228), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 39/69 (56%), Positives = 53/69 (76%), Gaps = 0/69 (0%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFPEK+QLK Y +E+AK+RDHR +G +L LF+F+ S G FWLP GAK+YN L
Sbjct 354 YGISFPEKEQLKMYKHRIEEAKQRDHRTIGTDLKLFYFDTVHSPGSCFWLPNGAKIYNRL 413
Query 61 IEFIREEYQ 69
+EF+R+ Y+
Sbjct 414 VEFMRDNYR 422
> pfa:PF11_0270 threonine --tRNA ligase, putative; K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=1013
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 42/69 (60%), Positives = 53/69 (76%), Gaps = 0/69 (0%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SF +K +L EYL LE+AK+RDHR +G L+LFFFE S G FWLP G+K+YN L
Sbjct 506 YGISFQKKSELVEYLKFLEEAKKRDHRNVGKILNLFFFEKETSPGSCFWLPHGSKIYNKL 565
Query 61 IEFIREEYQ 69
IEFIR+EY+
Sbjct 566 IEFIRKEYR 574
> tgo:TGME49_100260 threonyl-tRNA synthetase, putative (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=844
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 48/69 (69%), Positives = 57/69 (82%), Gaps = 0/69 (0%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YGVSFP+KK LK+YL LLE+AK+RDHR+LG NL LFFF+ VS G FWLP+GAKVYN L
Sbjct 372 YGVSFPDKKLLKDYLKLLEEAKKRDHRVLGQNLHLFFFDTNVSPGSCFWLPDGAKVYNKL 431
Query 61 IEFIREEYQ 69
F+REEY+
Sbjct 432 CMFMREEYR 440
> bbo:BBOV_IV008180 23.m05860; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=736
Score = 85.5 bits (210), Expect = 3e-17, Method: Composition-based stats.
Identities = 38/69 (55%), Positives = 49/69 (71%), Gaps = 0/69 (0%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP++ QLK Y +E AK DHRLLG L+LF F+ S G FWLP GAK+YN L
Sbjct 301 YGISFPKEAQLKAYERRIEDAKANDHRLLGTTLNLFHFDNIHSPGSCFWLPNGAKIYNRL 360
Query 61 IEFIREEYQ 69
++F+RE Y+
Sbjct 361 VDFMRENYR 369
> cel:C47D12.6 trs-1; Threonyl tRNA Synthetase family member (trs-1);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=725
Score = 82.0 bits (201), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 38/69 (55%), Positives = 51/69 (73%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ KQLKE+ L E+A +RDHR LG LFFF +S G AFW P+GA +YN L
Sbjct 297 YGISFPDSKQLKEWQKLQEEAAKRDHRKLGKEHDLFFFH-QLSPGSAFWYPKGAHIYNKL 355
Query 61 IEFIREEYQ 69
++FIR++Y+
Sbjct 356 VDFIRKQYR 364
> hsa:6897 TARS, MGC9344, ThrRS; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=723
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 37/69 (53%), Positives = 49/69 (71%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ K LKE+ E+AK RDHR +G + L+FF +S G F+LP+GA +YN L
Sbjct 298 YGISFPDPKMLKEWEKFQEEAKNRDHRKIGRDQELYFFH-ELSPGSCFFLPKGAYIYNAL 356
Query 61 IEFIREEYQ 69
IEFIR EY+
Sbjct 357 IEFIRSEYR 365
> xla:447636 tars, MGC86352; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=721
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 38/69 (55%), Positives = 48/69 (69%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ K LKE+ E+AK RDHR LG LFFF +S G F+LP+GA +YN L
Sbjct 296 YGISFPDPKMLKEWEKFQEEAKNRDHRKLGKEQELFFFH-ELSPGSCFFLPKGAYIYNQL 354
Query 61 IEFIREEYQ 69
I FIR+EY+
Sbjct 355 IGFIRQEYR 363
> mmu:272396 Tarsl2, A530046H20Rik, MGC31414; threonyl-tRNA synthetase-like
2 (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=790
Score = 74.7 bits (182), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 35/69 (50%), Positives = 47/69 (68%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ K +K++ E+AK RDHR +G LFFF +S G F+LP GA +YN L
Sbjct 365 YGISFPDSKMMKDWEKFQEEAKSRDHRKIGKEQELFFFH-DLSPGSCFFLPRGAFIYNAL 423
Query 61 IEFIREEYQ 69
++FIREEY
Sbjct 424 MDFIREEYH 432
> ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligase
(THRRS) (EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=709
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 36/68 (52%), Positives = 50/68 (73%), Gaps = 1/68 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+S+P++KQLK+YL LE+AK+ DHRLLG L FF +S G F+LP G +VYN L
Sbjct 292 YGISYPDQKQLKKYLQFLEEAKKYDHRLLGQKQEL-FFSHQLSPGSYFFLPLGTRVYNRL 350
Query 61 IEFIREEY 68
++FI+ +Y
Sbjct 351 MDFIKNQY 358
> mmu:110960 Tars, D15Wsu59e, ThrRS; threonyl-tRNA synthetase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=722
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/69 (52%), Positives = 49/69 (71%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ K LKE+ E+AK RDHR +G + L+FF +S G F+LP+GA +YN L
Sbjct 297 YGISFPDPKLLKEWEKFQEEAKNRDHRKIGRDQELYFFH-ELSPGSCFFLPKGAYIYNTL 355
Query 61 IEFIREEYQ 69
+EFIR EY+
Sbjct 356 MEFIRSEYR 364
> hsa:123283 TARSL2, FLJ25005; threonyl-tRNA synthetase-like 2
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=802
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 46/69 (66%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+ K ++++ E+AK RDHR +G LFFF +S G F+LP GA +YN L
Sbjct 377 YGISFPDNKMMRDWEKFQEEAKNRDHRKIGKEQELFFFH-DLSPGSCFFLPRGAFIYNTL 435
Query 61 IEFIREEYQ 69
+FIREEY
Sbjct 436 TDFIREEYH 444
> sce:YIL078W THS1; Ths1p (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=734
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 31/69 (44%), Positives = 44/69 (63%), Gaps = 1/69 (1%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG+SFP+KK + +L L +A RDHR +G LF F +S G FWLP G ++YN L
Sbjct 305 YGISFPDKKLMDAHLKFLAEASMRDHRKIGKEQELFLFN-EMSPGSCFWLPHGTRIYNTL 363
Query 61 IEFIREEYQ 69
++ +R EY+
Sbjct 364 VDLLRTEYR 372
> dre:100333765 Threonyl-tRNA synthetase, cytoplasmic-like
Length=529
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 30/68 (44%), Positives = 46/68 (67%), Gaps = 1/68 (1%)
Query 2 GVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGLI 61
GVSFP +++ +++ E+A+ RDHR +G LFFF VS G F++P+GA +YN L
Sbjct 108 GVSFPSEREREQWEKEQEEARRRDHRRIGKAQELFFFH-DVSPGSCFFMPKGAHIYNTLT 166
Query 62 EFIREEYQ 69
+FI+ EY+
Sbjct 167 DFIKSEYR 174
> mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.3); K01868
threonyl-tRNA synthetase [EC:6.1.1.3]
Length=697
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Query 2 GVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGLI 61
G+SFP+ + L+ + E A+ RDHR +G LFFF +S G F+LP G +VYN L+
Sbjct 283 GISFPKVELLRNWEARREAAELRDHRRIGKEQELFFFH-ELSPGSCFFLPRGTRVYNALV 341
Query 62 EFIREEY 68
FIR EY
Sbjct 342 AFIRAEY 348
> hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.3); K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=718
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 42/67 (62%), Gaps = 1/67 (1%)
Query 2 GVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGLI 61
G+SFP + L+ + E+A+ RDHR +G LFFF +S G F+LP G +VYN L+
Sbjct 278 GISFPTTELLRVWEAWREEAELRDHRRIGKEQELFFFH-ELSPGSCFFLPRGTRVYNALV 336
Query 62 EFIREEY 68
FIR EY
Sbjct 337 AFIRAEY 343
> ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding /
aminoacyl-tRNA ligase/ ligase, forming aminoacyl-tRNA and related
compounds / nucleotide binding / threonine-tRNA ligase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=650
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/58 (44%), Positives = 35/58 (60%), Gaps = 0/58 (0%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYN 58
YG ++ ++QLK YL E+AK RDHR +G +L LF + G FW P+GA V N
Sbjct 230 YGTAWESEEQLKAYLHFKEEAKRRDHRRIGQDLDLFSIQDEAGGGLVFWHPKGAIVRN 287
> eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=642
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 41/72 (56%), Gaps = 4/72 (5%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGL 60
YG ++ +KK L YL LE+A +RDHR +G L L+ + + G FW +G ++ L
Sbjct 219 YGTAWADKKALNAYLQRLEEAAKRDHRKIGKQLDLYHMQEE-APGMVFWHNDGWTIFREL 277
Query 61 IEFIR---EEYQ 69
F+R +EYQ
Sbjct 278 EVFVRSKLKEYQ 289
> dre:100330276 threonyl-tRNA synthetase-like
Length=380
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/30 (56%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 1 YGVSFPEKKQLKEYLDLLEKAKERDHRLLG 30
YG+SFP+ K LKE+ E+AK RDHR +G
Sbjct 98 YGISFPDSKMLKEWERFQEEAKNRDHRKIG 127
> sce:YKL194C MST1; Mitochondrial threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=462
Score = 35.8 bits (81), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Query 28 LLGNNLSLFFFEPTVSAGPAFWLPEGAKVYNGLIEFIR 65
++ LF +P +S G F+LP GAK++N LIEF++
Sbjct 41 MVSQRQDLFMTDP-LSPGSMFFLPNGAKIFNKLIEFMK 77
> cel:Y61A9LA.10 hypothetical protein; K14569 ribosome biogenesis
protein BMS1
Length=1055
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 19 EKAKERDHRLLGNNLSLFFFEPTVSAGPA 47
+K E++ R+ GNNL F F VSAG A
Sbjct 25 KKRNEKEPRVKGNNLKAFTFHSAVSAGKA 53
> tgo:TGME49_099260 hypothetical protein
Length=131
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 21/47 (44%), Gaps = 0/47 (0%)
Query 5 FPEKKQLKEYLDLLEKAKERDHRLLGNNLSLFFFEPTVSAGPAFWLP 51
F Q +LD + ++ E+D + G N F EP GP LP
Sbjct 68 FTASVQQSVHLDFVLESAEKDKIIRGTNAQAFGVEPRYDCGPFAHLP 114
> cel:F22B7.6 polk-1; POLK (DNA polymerase kappa) homolog family
member (polk-1); K03511 DNA polymerase kappa subunit [EC:2.7.7.7]
Length=596
Score = 27.7 bits (60), Expect = 9.6, Method: Composition-based stats.
Identities = 14/25 (56%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 15 LDLLEKAKERDHRLLGNNLSLFFFE 39
L+LLEK K ++ RLLG LS FE
Sbjct 452 LELLEKEKGKEIRLLGVRLSQLIFE 476
Lambda K H
0.319 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2029389012
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40