bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5400_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
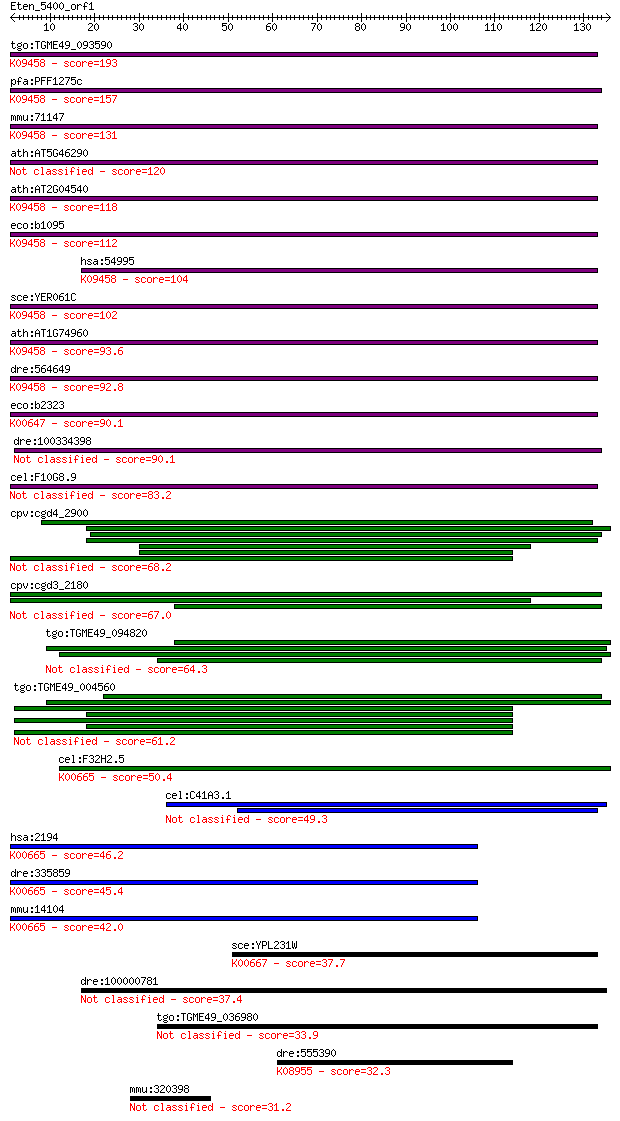
tgo:TGME49_093590 3-oxoacyl-[acyl-carrier-protein] synthase, p... 193 1e-49
pfa:PFF1275c fabB, fabF; 3-oxoacyl-acyl-carrier protein syntha... 157 7e-39
mmu:71147 Oxsm, 4933425A18Rik, C80494; 3-oxoacyl-ACP synthase,... 131 5e-31
ath:AT5G46290 KAS_I; KAS I (3-KETOACYL-ACYL CARRIER PROTEIN SY... 120 8e-28
ath:AT2G04540 3-oxoacyl-(acyl-carrier-protein) synthase II, pu... 118 6e-27
eco:b1095 fabF, cvc, ECK1081, fabJ, JW1081, vtr; 3-oxoacyl-[ac... 112 2e-25
hsa:54995 OXSM, FASN2D, FLJ20604, KASI, KS; 3-oxoacyl-ACP synt... 104 1e-22
sce:YER061C CEM1; Cem1p (EC:2.3.1.41); K09458 3-oxoacyl-[acyl-... 102 4e-22
ath:AT1G74960 FAB1; FAB1 (FATTY ACID BIOSYNTHESIS 1); 3-oxoacy... 93.6 2e-19
dre:564649 similar to 3-oxoacyl-[acyl-carrier-protein] synthas... 92.8 3e-19
eco:b2323 fabB, ECK2317, fabC, JW2320; 3-oxoacyl-[acyl-carrier... 90.1 2e-18
dre:100334398 plastid 3-keto-acyl-ACP synthase I-like 90.1 2e-18
cel:F10G8.9 hypothetical protein 83.2 2e-16
cpv:cgd4_2900 polyketide synthase 68.2 7e-12
cpv:cgd3_2180 type I fatty acid synthase 67.0 1e-11
tgo:TGME49_094820 type I fatty acid synthase, putative (EC:6.2... 64.3 9e-11
tgo:TGME49_004560 type I fatty acid synthase, putative (EC:5.1... 61.2 9e-10
cel:F32H2.5 fasn-1; Fatty Acid SyNthase family member (fasn-1)... 50.4 2e-06
cel:C41A3.1 hypothetical protein 49.3 4e-06
hsa:2194 FASN, FAS, MGC14367, MGC15706, OA-519, SDR27X1; fatty... 46.2 3e-05
dre:335859 fasn, fj34h12, wu:fj34h12; fatty acid synthase; K00... 45.4 5e-05
mmu:14104 Fasn, A630082H08Rik, FAS; fatty acid synthase (EC:2.... 42.0 6e-04
sce:YPL231W FAS2; Alpha subunit of fatty acid synthetase, whic... 37.7 0.011
dre:100000781 si:dkey-61p9.11 37.4 0.012
tgo:TGME49_036980 type I fatty acid synthase, putative (EC:6.2... 33.9 0.14
dre:555390 fc01d11; wu:fc01d11; K08955 ATP-dependent metallopr... 32.3 0.47
mmu:320398 Lrig3, 9030421L11Rik, 9130004I02Rik, 9430095K15Rik,... 31.2 1.0
> tgo:TGME49_093590 3-oxoacyl-[acyl-carrier-protein] synthase,
putative (EC:2.3.1.41); K09458 3-oxoacyl-[acyl-carrier-protein]
synthase II [EC:2.3.1.179]
Length=551
Score = 193 bits (491), Expect = 1e-49, Method: Compositional matrix adjust.
Identities = 91/133 (68%), Positives = 110/133 (82%), Gaps = 1/133 (0%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
++V ETEEHAVRRGAP +YG++AGYGA+CDAHH+TAP+P+G GL+RC+E A+ DA +
Sbjct 368 IMVFETEEHAVRRGAPKIYGEVAGYGASCDAHHITAPAPDGSGLSRCLENAIADANIDKR 427
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFG-PHATKLLISSTKPMTGHCLGGTAGIETVVAA 119
D+GYINAHGTST ND+IETKA KKVFG A +LLISSTK MTGHCLG T GIE V+A
Sbjct 428 DIGYINAHGTSTPMNDRIETKAFKKVFGDEQARQLLISSTKSMTGHCLGATGGIEACVSA 487
Query 120 KVLQTGDVPATLN 132
KVL+TG VP T+N
Sbjct 488 KVLETGIVPPTIN 500
> pfa:PFF1275c fabB, fabF; 3-oxoacyl-acyl-carrier protein synthase
I/II (EC:2.3.1.41); K09458 3-oxoacyl-[acyl-carrier-protein]
synthase II [EC:2.3.1.179]
Length=473
Score = 157 bits (398), Expect = 7e-39, Method: Composition-based stats.
Identities = 72/133 (54%), Positives = 94/133 (70%), Gaps = 1/133 (0%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
+++LE+ EHA++R AP +YG++ Y + CDA+H+TAP P G GL + ALK+A + +
Sbjct 295 ILILESYEHAIKRNAP-IYGEIISYSSECDAYHITAPEPNGKGLTNSIHKALKNANININ 353
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAK 120
DV YINAHGTST NDKIETK K VF HA KL ISSTK MTGHC+G IE++V K
Sbjct 354 DVKYINAHGTSTNLNDKIETKVFKNVFKDHAYKLYISSTKSMTGHCIGAAGAIESIVCLK 413
Query 121 VLQTGDVPATLNY 133
+QT +P T+NY
Sbjct 414 TMQTNIIPPTINY 426
> mmu:71147 Oxsm, 4933425A18Rik, C80494; 3-oxoacyl-ACP synthase,
mitochondrial (EC:2.3.1.41); K09458 3-oxoacyl-[acyl-carrier-protein]
synthase II [EC:2.3.1.179]
Length=459
Score = 131 bits (330), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 67/132 (50%), Positives = 85/132 (64%), Gaps = 1/132 (0%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
V+VLE EHAV+RGA +Y ++ GYG + DA H+TAP PEG+G RCM A+KDAGV+P+
Sbjct 282 VLVLEEHEHAVQRGA-RIYAEILGYGLSGDAGHITAPDPEGEGALRCMAAAVKDAGVSPE 340
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAK 120
+ Y+NAH TST D E +AIK++F HA L ISSTK TGH LG +E A
Sbjct 341 QISYVNAHATSTPLGDAAENRAIKRLFRDHACALAISSTKGATGHLLGAAGAVEATFTAL 400
Query 121 VLQTGDVPATLN 132
+P TLN
Sbjct 401 ACYHQKLPPTLN 412
> ath:AT5G46290 KAS_I; KAS I (3-KETOACYL-ACYL CARRIER PROTEIN
SYNTHASE I); catalytic/ fatty-acid synthase/ transferase, transferring
acyl groups other than amino-acyl groups
Length=418
Score = 120 bits (302), Expect = 8e-28, Method: Composition-based stats.
Identities = 62/132 (46%), Positives = 86/132 (65%), Gaps = 2/132 (1%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
V+V+E+ EHA++RGAP V L G CDAHH+T P +G G++ C+E L+DAGV+P+
Sbjct 243 VLVMESLEHAMKRGAPIVAEYLGG-AVNCDAHHMTDPRADGLGVSSCIERCLEDAGVSPE 301
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAK 120
+V YINAH TST D E AIKKVF + + I++TK M GHCLG G+E + K
Sbjct 302 EVNYINAHATSTLAGDLAEINAIKKVF-KSTSGIKINATKSMIGHCLGAAGGLEAIATVK 360
Query 121 VLQTGDVPATLN 132
+ TG + ++N
Sbjct 361 AINTGWLHPSIN 372
> ath:AT2G04540 3-oxoacyl-(acyl-carrier-protein) synthase II,
putative (EC:2.3.1.179); K09458 3-oxoacyl-[acyl-carrier-protein]
synthase II [EC:2.3.1.179]
Length=461
Score = 118 bits (295), Expect = 6e-27, Method: Compositional matrix adjust.
Identities = 63/134 (47%), Positives = 80/134 (59%), Gaps = 3/134 (2%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
VIVLE EHA RRGA +Y +L GYG + DAHH+T P +G G M AL+ +G+ P+
Sbjct 284 VIVLEEYEHAKRRGA-KIYAELCGYGMSGDAHHITQPPEDGKGAVLAMTRALRQSGLCPN 342
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATK--LLISSTKPMTGHCLGGTAGIETVVA 118
+ Y+NAH TST D +E +AIK VF HAT L SSTK TGH LG +E + +
Sbjct 343 QIDYVNAHATSTPIGDAVEARAIKTVFSEHATSGTLAFSSTKGATGHLLGAAGAVEAIFS 402
Query 119 AKVLQTGDVPATLN 132
+ G P TLN
Sbjct 403 ILAIHHGVAPMTLN 416
> eco:b1095 fabF, cvc, ECK1081, fabJ, JW1081, vtr; 3-oxoacyl-[acyl-carrier-protein]
synthase II (EC:2.3.1.41); K09458 3-oxoacyl-[acyl-carrier-protein]
synthase II [EC:2.3.1.179]
Length=413
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 61/132 (46%), Positives = 88/132 (66%), Gaps = 1/132 (0%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
++VLE EHA +RGA +Y +L G+G + DA+H+T+P G G A M AL+DAG+
Sbjct 238 MLVLEEYEHAKKRGA-KIYAELVGFGMSSDAYHMTSPPENGAGAALAMANALRDAGIEAS 296
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAK 120
+GY+NAHGTST DK E +A+K +FG A+++L+SSTK MTGH LG +E++ +
Sbjct 297 QIGYVNAHGTSTPAGDKAEAQAVKTIFGEAASRVLVSSTKSMTGHLLGAAGAVESIYSIL 356
Query 121 VLQTGDVPATLN 132
L+ VP T+N
Sbjct 357 ALRDQAVPPTIN 368
> hsa:54995 OXSM, FASN2D, FLJ20604, KASI, KS; 3-oxoacyl-ACP synthase,
mitochondrial (EC:2.3.1.41); K09458 3-oxoacyl-[acyl-carrier-protein]
synthase II [EC:2.3.1.179]
Length=376
Score = 104 bits (259), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 56/116 (48%), Positives = 66/116 (56%), Gaps = 0/116 (0%)
Query 17 HVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYND 76
H G + A DA H+TAP PEG+G RCM ALKDAGV P+++ YINAH TST D
Sbjct 214 HAVGDSFRFIAHGDAGHITAPDPEGEGALRCMAAALKDAGVQPEEISYINAHATSTPLGD 273
Query 77 KIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAKVLQTGDVPATLN 132
E KAIK +F HA L +SSTK TGH LG +E +P TLN
Sbjct 274 AAENKAIKHLFKDHAYALAVSSTKGATGHLLGAAGAVEAAFTTLACYYQKLPPTLN 329
> sce:YER061C CEM1; Cem1p (EC:2.3.1.41); K09458 3-oxoacyl-[acyl-carrier-protein]
synthase II [EC:2.3.1.179]
Length=442
Score = 102 bits (254), Expect = 4e-22, Method: Composition-based stats.
Identities = 56/135 (41%), Positives = 79/135 (58%), Gaps = 4/135 (2%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
+IV+E+ EHA +R A ++ +L GYG + DA H+T+P +G+G R +E+ALK A + P
Sbjct 256 MIVMESLEHAQKRNA-NIISELVGYGLSSDACHITSPPADGNGAKRAIEMALKMARLEPT 314
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATK---LLISSTKPMTGHCLGGTAGIETVV 117
DV Y+NAH TST DK E A+ P +K L ISS K GH LG +E++
Sbjct 315 DVDYVNAHATSTLLGDKAECLAVASALLPGRSKSKPLYISSNKGAIGHLLGARGAVESIF 374
Query 118 AAKVLQTGDVPATLN 132
L+ +P TLN
Sbjct 375 TICSLKDDKMPHTLN 389
> ath:AT1G74960 FAB1; FAB1 (FATTY ACID BIOSYNTHESIS 1); 3-oxoacyl-[acyl-carrier-protein]
synthase/ fatty-acid synthase (EC:2.3.1.41);
K09458 3-oxoacyl-[acyl-carrier-protein] synthase
II [EC:2.3.1.179]
Length=541
Score = 93.6 bits (231), Expect = 2e-19, Method: Composition-based stats.
Identities = 55/132 (41%), Positives = 80/132 (60%), Gaps = 2/132 (1%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
V++LE EHA +RGA +Y + G TCDA+H+T P P+G G+ C+E AL AG++ +
Sbjct 366 VLLLEELEHAKKRGAT-IYAEFLGGSFTCDAYHMTEPHPDGAGVILCIERALASAGISKE 424
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAK 120
+ YINAH TST D E +A+ FG + +L ++STK M GH LG +E V +
Sbjct 425 QINYINAHATSTHAGDIKEYQALAHCFGQNP-ELKVNSTKSMIGHLLGAAGAVEAVATVQ 483
Query 121 VLQTGDVPATLN 132
++TG V +N
Sbjct 484 AIRTGWVHPNIN 495
> dre:564649 similar to 3-oxoacyl-[acyl-carrier-protein] synthase,
mitochondrial precursor (Beta-ketoacyl-ACP synthase); K09458
3-oxoacyl-[acyl-carrier-protein] synthase II [EC:2.3.1.179]
Length=451
Score = 92.8 bits (229), Expect = 3e-19, Method: Composition-based stats.
Identities = 57/132 (43%), Positives = 71/132 (53%), Gaps = 4/132 (3%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
V+VLE A RRGA VY A +H P+ +G+ RCM AL++AG++P
Sbjct 277 VLVLEEFSRAQRRGA-RVY---AXXXXXXXSHLYLLPAAVSNGVFRCMSAALRNAGLSPS 332
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAK 120
DV YINAH TST D E AIK++F P L +SSTK TGH LG +ET A
Sbjct 333 DVSYINAHATSTPLGDAAENAAIKRLFEPSVLSLAVSSTKGATGHLLGAAGALETAFTAL 392
Query 121 VLQTGDVPATLN 132
+P TLN
Sbjct 393 ACHHAILPPTLN 404
> eco:b2323 fabB, ECK2317, fabC, JW2320; 3-oxoacyl-[acyl-carrier-protein]
synthase I (EC:2.3.1.41); K00647 3-oxoacyl-[acyl-carrier-protein]
synthase I [EC:2.3.1.41]
Length=406
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 54/132 (40%), Positives = 79/132 (59%), Gaps = 8/132 (6%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
++V+E EHA+ RGA H+Y ++ GYGAT D + APS G+G RCM++A+ D
Sbjct 237 MVVVEELEHALARGA-HIYAEIVGYGATSDGADMVAPS--GEGAVRCMKMAMHGVDTPID 293
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAK 120
Y+N+HGTST D E AI++VFG + IS+TK MTGH LG E + +
Sbjct 294 ---YLNSHGTSTPVGDVKELAAIREVFGDKSPA--ISATKAMTGHSLGAAGVQEAIYSLL 348
Query 121 VLQTGDVPATLN 132
+L+ G + ++N
Sbjct 349 MLEHGFIAPSIN 360
> dre:100334398 plastid 3-keto-acyl-ACP synthase I-like
Length=429
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 57/133 (42%), Positives = 74/133 (55%), Gaps = 2/133 (1%)
Query 2 IVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPDD 61
+VLE+ E AV RGA V G L G G D H T SP+G + AL+DAG+
Sbjct 253 LVLESLESAVARGA-KVLGILKGCGEKADHFHRTRSSPDGGPAIATIRAALEDAGMDESG 311
Query 62 VGYINAHGTSTFYNDKIETKAIKKVFGPHATK-LLISSTKPMTGHCLGGTAGIETVVAAK 120
+GYINAHGTST NDK+E A+ VFG + SS K M GH L +E V + +
Sbjct 312 IGYINAHGTSTPENDKMEYGAMLAVFGDRLKDGIPCSSNKSMIGHTLTAAGAVEAVFSIQ 371
Query 121 VLQTGDVPATLNY 133
+ TG +P T+N+
Sbjct 372 TMLTGTLPPTINH 384
> cel:F10G8.9 hypothetical protein
Length=403
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/132 (32%), Positives = 69/132 (52%), Gaps = 3/132 (2%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
++ +E E A RGA + ++ GYG + D +H++ P P G M A+ +A + P
Sbjct 226 LVFMERLEDAQARGA-QILAEVVGYGISSDCYHISTPDPSAIGAVLSMNRAIGNAHLEPK 284
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAK 120
D+GY+NAH TST D +E +A+++VF + +SS K GH LG +E +
Sbjct 285 DIGYVNAHATSTPNGDSVEAEAVRQVFPEQ--NIAVSSVKGHIGHLLGAAGSVEAIATIF 342
Query 121 VLQTGDVPATLN 132
+ +PA N
Sbjct 343 AMNDDVLPANRN 354
> cpv:cgd4_2900 polyketide synthase
Length=13413
Score = 68.2 bits (165), Expect = 7e-12, Method: Composition-based stats.
Identities = 39/125 (31%), Positives = 64/125 (51%), Gaps = 2/125 (1%)
Query 8 EHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINA 67
E+ ++ +YG++ G D + +P G R + A++D+ ++ +D+ YI +
Sbjct 3028 ENHLKSFGKKIYGRICGTAVNQDGKSASLTAPNGPSQKRVIINAIEDSCISNNDIFYIES 3087
Query 68 HGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAA-KVLQTGD 126
HGT T D IE AI VF A KL I + KP GH L G +G+ ++ A V+ G
Sbjct 3088 HGTGTPLGDPIEFGAISDVFKNRANKLYIGTLKPNIGH-LEGASGVAGILKAILVVNNGV 3146
Query 127 VPATL 131
+P +
Sbjct 3147 IPPNI 3151
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 39/124 (31%), Positives = 60/124 (48%), Gaps = 11/124 (8%)
Query 18 VYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDK 77
+YG + G D + +P G + ++ A++D+ + D++ I AHGT T D
Sbjct 11221 IYGFIKGSAVNQDGRSASLTAPNGPAQQKVIKSAIEDSYIKLDEISIIEAHGTGTQLGDP 11280
Query 78 IETKAIKKVF-----GPHATKLLISSTKPMTGHCLGGTAGIETVVAA-KVLQTGDVPATL 131
IE AIK VF GP + I++ K GH L G +GI V+ +L VP L
Sbjct 11281 IEFGAIKNVFNSRNRGP----IYITALKTNIGH-LEGASGIAGVIKMILMLNHSIVPKNL 11335
Query 132 NYST 135
++ T
Sbjct 11336 HFKT 11339
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 39/127 (30%), Positives = 58/127 (45%), Gaps = 13/127 (10%)
Query 19 YGQLAGYGATCDAHHVTAPSPEGDGLARCMEIAL---KDAGVAPDDVGYINAHGTSTFYN 75
+G++ G ++ +P G + + AL ++ + GYI HGT T
Sbjct 4746 WGRILGSSTNHSGRSISLTAPNGVAQQKVLSTALNIAREENKSDIRPGYIECHGTGTLLG 4805
Query 76 DKIETKAIKKVFGPH--------ATKLLISSTKPMTGHCLGGTAGIETVVA-AKVLQTGD 126
D +E AIK+V+ + L++S+ KP GH L G AGI + VL G
Sbjct 4806 DAVEFGAIKEVYKDMYKIIDSNIDSPLILSTVKPQIGH-LEGAAGIVGFIRLLVVLSFGA 4864
Query 127 VPATLNY 133
VPA LNY
Sbjct 4865 VPALLNY 4871
Score = 48.1 bits (113), Expect = 8e-06, Method: Composition-based stats.
Identities = 36/116 (31%), Positives = 54/116 (46%), Gaps = 2/116 (1%)
Query 18 VYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDK 77
V G L G D + +P + + AL + V ++V I +HGT T D
Sbjct 7931 VLGILKGTAVNQDGRTASLTAPSSKSQSDVISHALLRSEVQFNEVSMIESHGTGTQLGDP 7990
Query 78 IETKAIKKVFGPHATKLLISSTKPMTGHCLGGTAGIETVVAAKV-LQTGDVPATLN 132
IE AIK V+ + KL+I + K GH L +AGI ++ + L+ G P L+
Sbjct 7991 IEIDAIKTVYQLSSKKLVIGALKTNIGH-LEASAGIAGLIKLLLSLRNGISPPNLH 8045
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 30/100 (30%), Positives = 44/100 (44%), Gaps = 13/100 (13%)
Query 30 DAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFGP 89
D T +P G ++ A++ A ++P ++ YI HGT T D IE AIK VF
Sbjct 9468 DGKSATITAPNGPSQQFVIKHAIEKANLSPSNITYIETHGTGTKLGDPIEYNAIKSVFNL 9527
Query 90 HA------------TKLLISSTKPMTGHCLGGTAGIETVV 117
+ L + + K GH L G AGI ++
Sbjct 9528 EDNTFDNNVCLLRYSPLYLGAIKANIGH-LEGAAGIAGLI 9566
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 29/94 (30%), Positives = 41/94 (43%), Gaps = 11/94 (11%)
Query 30 DAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFGP 89
D T +P+G + L +G+ P ++ YI HGT T D IE AI VF
Sbjct 1102 DGKTTTLTAPKGPSQVSAIRNCLGISGMHPSEINYIECHGTGTPLGDPIEAGAIMSVFKT 1161
Query 90 H----------ATKLLISSTKPMTGHCLGGTAGI 113
H + ++I + K GH L G AG+
Sbjct 1162 HNEYQYNVTHMNSNIVIGAFKSNIGH-LEGAAGL 1194
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 29/116 (25%), Positives = 48/116 (41%), Gaps = 5/116 (4%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKD-AGVAP 59
+++L +E R V ++G D T +P + +C+ +AL + P
Sbjct 6679 IVILPKKE--CERKNKKVLAYISGSSVNQDGKSATLTAPSKNSQVKCINLALSSMEHIHP 6736
Query 60 DDVGYINAHGTSTFYNDKIETKAIKKVF--GPHATKLLISSTKPMTGHCLGGTAGI 113
V + AHGT T D IE A+ + + +KL+I S K H + I
Sbjct 6737 SMVTIVEAHGTGTLIGDSIEVSALYEAYCSSERKSKLMIGSGKSNFAHTFAASGII 6792
> cpv:cgd3_2180 type I fatty acid synthase
Length=8243
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 45/138 (32%), Positives = 67/138 (48%), Gaps = 9/138 (6%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
V+VL+T E+A +Y + G + + +P G + AL+ AG+ P+
Sbjct 1126 VLVLKTMENA--NNGEKIYSVIKGSSVNHNGRSASLTAPNGISQQNVINSALEYAGIRPN 1183
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFG---PHATKLLISSTKPMTGHCLG--GTAGIET 115
D+GYI AHGT T D IE A+K +F L+I + K GH G G AGI
Sbjct 1184 DIGYIEAHGTGTSLGDPIEVSALKSIFSRTRDSGKPLIIGAVKTNIGHLEGAAGMAGIFK 1243
Query 116 VVAAKVLQTGDVPATLNY 133
V+ + + T VP L++
Sbjct 1244 VILSLINDT--VPPNLHF 1259
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 36/122 (29%), Positives = 61/122 (50%), Gaps = 8/122 (6%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPD 60
+++L+ E ++ P + G++ G+G + +P G +++AL + +
Sbjct 3280 IVILKQSEQK-KQNTP-ILGRIKGWGCNHVGRSASLTAPNGPAQTCVIKMALNQSKLNSS 3337
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHA-----TKLLISSTKPMTGHCLGGTAGIET 115
D+ YI HGT T D IE A+K VFG +A T L++ + K GH L G AGI
Sbjct 3338 DIDYIETHGTGTALGDPIELGALKSVFGRNADYKRSTPLVLGALKSNIGH-LEGAAGIAG 3396
Query 116 VV 117
++
Sbjct 3397 LI 3398
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 32/102 (31%), Positives = 53/102 (51%), Gaps = 7/102 (6%)
Query 38 SPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFG-----PHAT 92
+P G ++ AL++A + P+D+ Y+ +HGT T D IE A+K VFG +
Sbjct 5541 APNGPSQTEVIQNALRNARLTPNDIDYLESHGTGTPLGDPIEFGALKTVFGNKKDIKRSQ 5600
Query 93 KLLISSTKPMTGHCLGGTAGIETVVA-AKVLQTGDVPATLNY 133
L++ + K GH L G AG+ ++ VL+ P L++
Sbjct 5601 PLILGALKTNIGH-LEGAAGVSGLLKLILVLKNRIAPKILHF 5641
> tgo:TGME49_094820 type I fatty acid synthase, putative (EC:6.2.1.3
1.1.1.1 1.2.1.31 1.1.1.9 2.3.1.94 1.1.1.36 2.3.1.161
2.3.1.86 2.3.1.39 2.3.1.16 2.8.2.20 1.6.5.5 1.1.1.14)
Length=10021
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 37/102 (36%), Positives = 58/102 (56%), Gaps = 5/102 (4%)
Query 38 SPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFGPHATK---L 94
+P G A + +AL+ AG+ P+D+ ++ +HGT T D IE A++ VF P K L
Sbjct 3353 APNGPSQAEVIRMALRRAGMKPNDLKFMESHGTGTSLGDPIEVNAVRTVFAPGREKTKPL 3412
Query 95 LISSTKPMTGHCLGGTAGIETVV-AAKVLQTGDVPATLNYST 135
LI + K GH L G++GI ++ A VL ++P L++ T
Sbjct 3413 LIGAVKTNIGH-LEGSSGIAGLIKACLVLSRREIPPNLHFKT 3453
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/105 (35%), Positives = 54/105 (51%), Gaps = 8/105 (7%)
Query 34 VTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFG----P 89
+TAPS G R + AL+ A + P D+ ++ HGT T D IE A+K VFG
Sbjct 1183 LTAPS--GPSQQRVIRTALRQAELTPGDIYFVETHGTGTSLGDPIEVGALKSVFGASRDA 1240
Query 90 HATKLLISSTKPMTGHCLGGTAGIETVVAAKV-LQTGDVPATLNY 133
L++ + K GH L G AGI V+ A + L+ +P L++
Sbjct 1241 KTKPLILGAAKTNIGH-LEGAAGIAGVLKAMLALRYKHIPPNLHF 1284
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 38/128 (29%), Positives = 61/128 (47%), Gaps = 5/128 (3%)
Query 12 RRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTS 71
+R + G + G D + +P G + AL+ GV P DV + +HGT
Sbjct 7251 QREGKRILGIVRGTAVNHDGRSASLTAPNGPAQQDVIRSALQIGGVDPLDVALVESHGTG 7310
Query 72 TFYNDKIETKAIKKVFG---PHATKLLISSTKPMTGHCLGGTAGIETVVAAKV-LQTGDV 127
T D IE AIK V+G + L++ + K GH L G++GI ++ + L+ +V
Sbjct 7311 TALGDPIEMGAIKAVYGAGRSADSPLVVGALKSYIGH-LEGSSGIAGILKVLLCLRHHEV 7369
Query 128 PATLNYST 135
P L++ T
Sbjct 7370 PPNLHFDT 7377
Score = 52.0 bits (123), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 38/130 (29%), Positives = 58/130 (44%), Gaps = 5/130 (3%)
Query 9 HAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAH 68
A R+ + + G D + +P G + AL+ GV P D+ + +H
Sbjct 5497 QAARKDDSTILAVVRGSAINHDGRSASLTAPNGPAQQAVIRAALQSGGVKPADITVLESH 5556
Query 69 GTSTFYNDKIETKAIKKVFGPHA---TKLLISSTKPMTGHCLGGTAGIETVVAAKV-LQT 124
GT T D IE AIK VF H + I + K GH L G AGI + + + L+
Sbjct 5557 GTGTQLGDPIEVGAIKAVFMHHRGADNPIYIGALKTNIGH-LEGAAGIASFIKLVLCLRR 5615
Query 125 GDVPATLNYS 134
++PA L++
Sbjct 5616 RELPANLHFQ 5625
> tgo:TGME49_004560 type I fatty acid synthase, putative (EC:5.1.1.11
1.2.1.31 2.3.1.161 2.3.1.86 2.3.1.39 2.3.1.94 2.3.1.41)
Length=6350
Score = 61.2 bits (147), Expect = 9e-10, Method: Composition-based stats.
Identities = 39/115 (33%), Positives = 55/115 (47%), Gaps = 3/115 (2%)
Query 22 LAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDKIETK 81
+ G+G + V+ +P + +E L+ A V P V Y+ AHGT T D IE +
Sbjct 274 IRGHGTNHNGRSVSVTAPSDVAQKKLLEGVLRTASVEPQQVAYLEAHGTGTKLGDTIEFR 333
Query 82 AIKKVFGPHAT---KLLISSTKPMTGHCLGGTAGIETVVAAKVLQTGDVPATLNY 133
AIK VF + LLI S K GH G + + A VL +VP TL++
Sbjct 334 AIKSVFSTSRSVDNPLLIGSGKSNVGHLEGCAGAVGLLKALLVLYFREVPPTLHF 388
Score = 49.7 bits (117), Expect = 3e-06, Method: Composition-based stats.
Identities = 43/132 (32%), Positives = 59/132 (44%), Gaps = 17/132 (12%)
Query 9 HAVRRGAPHVYGQLAGYGATCD-AHHVTAPSPEGDGLARCMEIALKDAGVAPDDVGYINA 67
HAV RG+ ATC SP L R + ++L+DA AP V Y A
Sbjct 5593 HAVVRGS-----------ATCHYGRSSQITSPNTRALTRVLRLSLQDASTAPSLVRYYEA 5641
Query 68 HGTSTFYNDKIETKAIKKVFGP---HATKLLISSTKPMTGHCLGGTAGIETVVAAKV-LQ 123
HGT+T D IE A+K VF A L + + GH L AGI + + L+
Sbjct 5642 HGTATVLGDVIEMSAVKDVFQTGRNAAAPLHMGTVHNNIGH-LDAAAGIVAFLKTVLCLK 5700
Query 124 TGDVPATLNYST 135
VPA +++ +
Sbjct 5701 HRFVPANIHFKS 5712
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 35/116 (30%), Positives = 50/116 (43%), Gaps = 7/116 (6%)
Query 2 IVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGV-APD 60
IV+E + RR PH ++ + SP G + AL+ A V +P
Sbjct 3485 IVIERDNERSRRPRPH--ARILSVTTNHVGRSASITSPNGPAQQAVIRAALRSANVNSPL 3542
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHA---TKLLISSTKPMTGHCLGGTAGI 113
V + HGT T D IE A++ V+G T L++ + K GH G AGI
Sbjct 3543 SVAVVETHGTGTSLGDPIEIGALQAVYGQGTSADTPLVLGALKSRIGHT-EGAAGI 3597
Score = 42.7 bits (99), Expect = 3e-04, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 45/100 (45%), Gaps = 5/100 (5%)
Query 18 VYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGV-APDDVGYINAHGTSTFYND 76
+Y LAG + + +P G + AL+ A V +P V + HGT T D
Sbjct 2975 IYAALAGTASNHVGRSASLTAPNGPAQQAVIRAALRSANVNSPLSVAVVETHGTGTSLGD 3034
Query 77 KIETKAIKKVFGPHA---TKLLISSTKPMTGHCLGGTAGI 113
IE A++ V+G T L++ + K GH G AGI
Sbjct 3035 PIEIGALQAVYGQGTSADTPLVLGALKSRIGHT-EGAAGI 3073
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 36/116 (31%), Positives = 50/116 (43%), Gaps = 6/116 (5%)
Query 2 IVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGV-APD 60
++LE RR YG+L G + SP G + AL+ A V +P
Sbjct 4069 LLLERTATPTRRNITP-YGRLLGTANNHVGRSASITSPNGPAQQAVIRAALRSANVNSPL 4127
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHA---TKLLISSTKPMTGHCLGGTAGI 113
V + HGT T D IE A++ V+G T L++ + K GH G AGI
Sbjct 4128 SVAVVETHGTGTSLGDPIEIGALQAVYGQGTSADTPLVLGALKSRIGHT-EGAAGI 4182
Score = 33.1 bits (74), Expect = 0.24, Method: Composition-based stats.
Identities = 27/100 (27%), Positives = 41/100 (41%), Gaps = 5/100 (5%)
Query 18 VYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVAP-DDVGYINAHGTSTFYND 76
+Y + G + +P G M L+ A + D+ + HGT T D
Sbjct 2252 IYAWIRGAATNHVGRSASLTAPNGPAQTTLMLDTLRSARLQRVSDISVLETHGTGTSLGD 2311
Query 77 KIETKAIKKVFGPHA---TKLLISSTKPMTGHCLGGTAGI 113
IE A++ V+G T L++ + K GH G AGI
Sbjct 2312 PIEIGALQAVYGQGTSADTPLVLGALKSRIGHT-EGAAGI 2350
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 30/116 (25%), Positives = 45/116 (38%), Gaps = 5/116 (4%)
Query 2 IVLETEEHAVRRGAPHVYGQLAGYGATCDAHHVTAPSPEGDGLARCMEIALKDAGVA-PD 60
+VL H + Y L G + +P G + +L+ A +
Sbjct 956 VVLLENTHDRQVSKNTTYAWLLGSANNHVGKSASLTAPNGPAQQAVILASLRSAHLQLTS 1015
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFGPHA---TKLLISSTKPMTGHCLGGTAGI 113
+ I HGT T D IE A++ V+G T L++ + K GH G AGI
Sbjct 1016 QIHLIETHGTGTSLGDPIEIGALQAVYGQGTSADTPLVLGALKSRIGHT-EGAAGI 1070
> cel:F32H2.5 fasn-1; Fatty Acid SyNthase family member (fasn-1);
K00665 fatty acid synthase, animal type [EC:2.3.1.85]
Length=2613
Score = 50.4 bits (119), Expect = 2e-06, Method: Composition-based stats.
Identities = 38/130 (29%), Positives = 62/130 (47%), Gaps = 10/130 (7%)
Query 12 RRGAPHVYGQLAGYGATCDAHH---VTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAH 68
++ A +Y + + D H +T PS G+ A+ ++ +AG+ P+ V Y+ AH
Sbjct 272 KKKAQRLYATVVHAKSNTDGHKEHGITFPS--GERQAQLLQEVYSEAGIDPNSVYYVEAH 329
Query 69 GTSTFYNDKIETKAIKKVFGPHAT-KLLISSTKPMTGHC--LGGTAGIETVVAAKVLQTG 125
GT T D E AI +VF T LLI S K GH G + ++ + ++
Sbjct 330 GTGTKVGDPQEANAICEVFCSKRTDSLLIGSVKSNMGHAEPASGVCSLTKILLS--IERQ 387
Query 126 DVPATLNYST 135
+P L+Y+T
Sbjct 388 LIPPNLHYNT 397
> cel:C41A3.1 hypothetical protein
Length=7829
Score = 49.3 bits (116), Expect = 4e-06, Method: Composition-based stats.
Identities = 29/100 (29%), Positives = 47/100 (47%), Gaps = 4/100 (4%)
Query 36 APSPEGDGLARCM-EIALKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFGPHATKL 94
AP+P G +CM ++ + + + ++ H T T D IE +++ + KL
Sbjct 5585 APNPAGQ--LKCMTDVLARFTNKEKERISFVECHATGTTLGDTIEMNSLRTAYS-FKNKL 5641
Query 95 LISSTKPMTGHCLGGTAGIETVVAAKVLQTGDVPATLNYS 134
I S K GH + V AK+LQTG +P +N+S
Sbjct 5642 AIGSCKANIGHAYAASGLAALVKCAKMLQTGIIPPQVNFS 5681
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 22/81 (27%), Positives = 43/81 (53%), Gaps = 8/81 (9%)
Query 52 LKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFGPHATKLLISSTKPMTGHCLGGTA 111
L++AG Y+ HGT+T D E+ A +K+ ++L++SS K GHC +
Sbjct 1087 LEEAG----SFSYVEGHGTATSAGDSAESMAYQKL----GSELIMSSVKAQFGHCEVASG 1138
Query 112 GIETVVAAKVLQTGDVPATLN 132
I+ + + + + G +P+ ++
Sbjct 1139 LIQLMKVSSIGKHGIIPSIVH 1159
> hsa:2194 FASN, FAS, MGC14367, MGC15706, OA-519, SDR27X1; fatty
acid synthase (EC:2.3.1.85 2.3.1.38 2.3.1.39 2.3.1.41 3.1.2.14
1.3.1.10 4.2.1.61 1.1.1.100); K00665 fatty acid synthase,
animal type [EC:2.3.1.85]
Length=2511
Score = 46.2 bits (108), Expect = 3e-05, Method: Composition-based stats.
Identities = 39/109 (35%), Positives = 47/109 (43%), Gaps = 10/109 (9%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDA---HHVTAPSPEGDGLARCMEIALKDAGV 57
V VL T++ RR VY + G D VT PS GD + + + AGV
Sbjct 229 VAVLLTKKSLARR----VYATILNAGTNTDGFKEQGVTFPS--GDIQEQLIRSLYQSAGV 282
Query 58 APDDVGYINAHGTSTFYNDKIETKAIKKVF-GPHATKLLISSTKPMTGH 105
AP+ YI AHGT T D E I + LLI STK GH
Sbjct 283 APESFEYIEAHGTGTKVGDPQELNGITRALCATRQEPLLIGSTKSNMGH 331
> dre:335859 fasn, fj34h12, wu:fj34h12; fatty acid synthase; K00665
fatty acid synthase, animal type [EC:2.3.1.85]
Length=2511
Score = 45.4 bits (106), Expect = 5e-05, Method: Composition-based stats.
Identities = 37/109 (33%), Positives = 50/109 (45%), Gaps = 10/109 (9%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHH---VTAPSPEGDGLARCMEIALKDAGV 57
V VL T++ +R +Y + G D + VT PS G+ R + ++A +
Sbjct 229 VAVLLTKKSMAKR----IYATVLNAGNNTDGYKEQGVTFPS--GEMQQRLVRSLYQEANI 282
Query 58 APDDVGYINAHGTSTFYNDKIETKAIKKVFGPHATK-LLISSTKPMTGH 105
+P+ V YI AHGT T D E I VF LLI STK GH
Sbjct 283 SPEQVEYIEAHGTGTKVGDPQEVNGIVSVFCQSPRDPLLIGSTKSNMGH 331
> mmu:14104 Fasn, A630082H08Rik, FAS; fatty acid synthase (EC:2.3.1.85
2.3.1.38 2.3.1.39 2.3.1.41 3.1.2.14 1.3.1.10 4.2.1.61
1.1.1.100); K00665 fatty acid synthase, animal type [EC:2.3.1.85]
Length=2504
Score = 42.0 bits (97), Expect = 6e-04, Method: Composition-based stats.
Identities = 38/109 (34%), Positives = 47/109 (43%), Gaps = 10/109 (9%)
Query 1 VIVLETEEHAVRRGAPHVYGQLAGYGATCDAHH---VTAPSPEGDGLARCMEIALKDAGV 57
V VL T++ RR VY + G D VT PS E C + AG+
Sbjct 229 VAVLLTKKSLARR----VYATILNAGTNTDGSKEQGVTFPSGEVQEQLICS--LYQPAGL 282
Query 58 APDDVGYINAHGTSTFYNDKIETKAI-KKVFGPHATKLLISSTKPMTGH 105
AP+ + YI AHGT T D E I + + LLI STK GH
Sbjct 283 APESLEYIEAHGTGTKVGDPQELNGITRSLCAFRQAPLLIGSTKSNMGH 331
> sce:YPL231W FAS2; Alpha subunit of fatty acid synthetase, which
catalyzes the synthesis of long-chain saturated fatty acids;
contains the acyl-carrier protein domain and beta-ketoacyl
reductase, beta-ketoacyl synthase and self-pantetheinylation
activities (EC:2.3.1.86 2.3.1.41 1.1.1.100); K00667 fatty
acid synthase subunit alpha, fungi type [EC:2.3.1.86]
Length=1887
Score = 37.7 bits (86), Expect = 0.011, Method: Composition-based stats.
Identities = 26/86 (30%), Positives = 39/86 (45%), Gaps = 4/86 (4%)
Query 51 ALKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFG----PHATKLLISSTKPMTGHC 106
AL G+ DD+G + HGTST NDK E+ I ++ ++ K +TGH
Sbjct 1525 ALATYGLTIDDLGVASFHGTSTKANDKNESATINEMMKHLGRSEGNPVIGVFQKFLTGHP 1584
Query 107 LGGTAGIETVVAAKVLQTGDVPATLN 132
G A ++L +G +P N
Sbjct 1585 KGAAGAWMMNGALQILNSGIIPGNRN 1610
> dre:100000781 si:dkey-61p9.11
Length=2105
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 37/129 (28%), Positives = 55/129 (42%), Gaps = 18/129 (13%)
Query 17 HVYGQLAGYGATCDAHHV---TAPSPEGDGLARCMEIALKDAGVAPD--DVGYINAHGTS 71
H++G + D H V T PS + + E+ K D V YI AHGT
Sbjct 260 HIWGVIGKTAVNQDGHSVSPITKPS-----MVQQEELLRKIYSTEADLCSVQYIEAHGTG 314
Query 72 TFYNDKIETKAIKKVFGPHATK----LLISSTKPMTGH--CLGGTAGIETVVAAKVLQTG 125
T D +E ++ KV ++ L+I S K GH G AG+ V+ ++Q
Sbjct 315 TPVGDPVEAGSMSKVIAKERSQKSGPLIIGSVKSNIGHTESAAGVAGLIKVLL--MMQHE 372
Query 126 DVPATLNYS 134
+ +L YS
Sbjct 373 TIVPSLFYS 381
> tgo:TGME49_036980 type I fatty acid synthase, putative (EC:6.2.1.3
2.3.1.94)
Length=1609
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 30/103 (29%), Positives = 40/103 (38%), Gaps = 6/103 (5%)
Query 34 VTAPSPEGDGLARCMEIALKDAGVAPDDVGYINAHGTSTFYNDKIETKAIKKVFGP---- 89
VTAPS L A A V P V + HGT T D IE A+ F P
Sbjct 1087 VTAPSGSSQELLYKQTFAC--AQVPPRAVDLVETHGTGTRLGDPIEISALCNSFAPDMET 1144
Query 90 HATKLLISSTKPMTGHCLGGTAGIETVVAAKVLQTGDVPATLN 132
+L++ + K GH G + + + L VP L+
Sbjct 1145 RKHRLVLGALKTNLGHLEGASGLAGLLKSVLCLNRQRVPPNLH 1187
> dre:555390 fc01d11; wu:fc01d11; K08955 ATP-dependent metalloprotease
[EC:3.4.24.-]
Length=2095
Score = 32.3 bits (72), Expect = 0.47, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 27/59 (45%), Gaps = 6/59 (10%)
Query 61 DVGYINAHGTSTFYNDKIETKAIKKVFG----PHATKLLISSTKPMTGH--CLGGTAGI 113
+V Y+ AHGT T D +E ++ +V P L + S K GH G AG+
Sbjct 301 NVQYVEAHGTGTPAGDPVEAGSLSRVIAKTRPPSLGPLFVGSVKSNIGHTESAAGVAGL 359
> mmu:320398 Lrig3, 9030421L11Rik, 9130004I02Rik, 9430095K15Rik,
mKIAA3016; leucine-rich repeats and immunoglobulin-like domains
3
Length=1117
Score = 31.2 bits (69), Expect = 1.0, Method: Composition-based stats.
Identities = 12/18 (66%), Positives = 15/18 (83%), Gaps = 0/18 (0%)
Query 28 TCDAHHVTAPSPEGDGLA 45
TCD+ H+TAPS +GDG A
Sbjct 789 TCDSPHMTAPSLDGDGWA 806
Lambda K H
0.316 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2296762580
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40