bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5381_orf1
Length=69
Score E
Sequences producing significant alignments: (Bits) Value
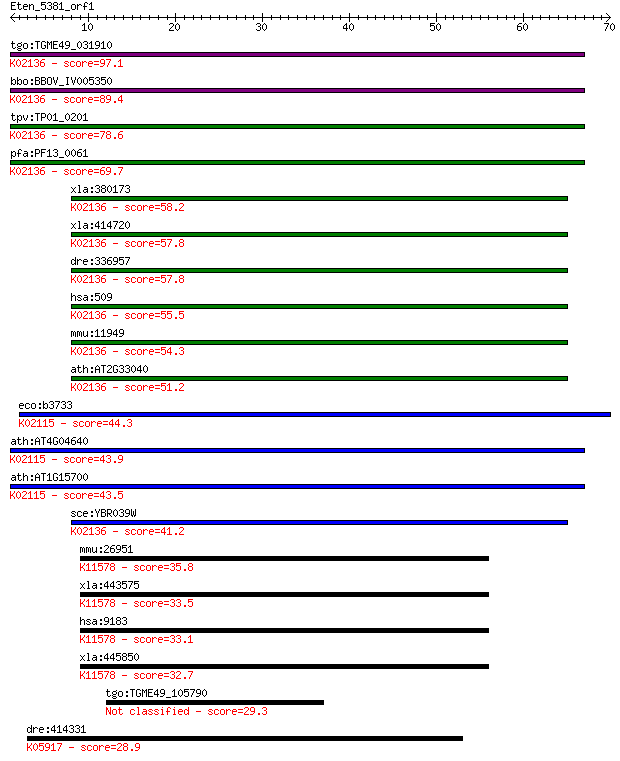
tgo:TGME49_031910 ATP synthase gamma chain, putative (EC:3.6.3... 97.1 1e-20
bbo:BBOV_IV005350 23.m06133; ATP synthase F1 gamma subunit (EC... 89.4 3e-18
tpv:TP01_0201 ATP synthase F1 subunit gamma; K02136 F-type H+-... 78.6 5e-15
pfa:PF13_0061 ATP synthase gamma chain, mitochondrial precurso... 69.7 2e-12
xla:380173 atp5c1, MGC53337; ATP synthase, H+ transporting, mi... 58.2 6e-09
xla:414720 hypothetical protein MGC82276; K02136 F-type H+-tra... 57.8 9e-09
dre:336957 atp5c1, wu:fk34c05, zgc:73209; ATP synthase, H+ tra... 57.8 9e-09
hsa:509 ATP5C1, ATP5C, ATP5CL1; ATP synthase, H+ transporting,... 55.5 4e-08
mmu:11949 Atp5c1, 1700094F02Rik; ATP synthase, H+ transporting... 54.3 8e-08
ath:AT2G33040 ATP synthase gamma chain, mitochondrial (ATPC); ... 51.2 8e-07
eco:b3733 atpG, ECK3726, JW3711, papC, uncG; F1 sector of memb... 44.3 1e-04
ath:AT4G04640 ATPC1; ATPC1; enzyme regulator; K02115 F-type H+... 43.9 1e-04
ath:AT1G15700 ATPC2; ATPC2; enzyme regulator; K02115 F-type H+... 43.5 2e-04
sce:YBR039W ATP3; Atp3p (EC:3.6.3.14); K02136 F-type H+-transp... 41.2 7e-04
mmu:26951 Zw10, 6330566F14Rik, MmZw10; ZW10 homolog (Drosophil... 35.8 0.038
xla:443575 zw10, MGC181630; centromere/kinetochore protein zw1... 33.5 0.17
hsa:9183 ZW10, HZW10, KNTC1AP, MGC149821; ZW10, kinetochore as... 33.1 0.22
xla:445850 zw10, MGC114854, xzw10; ZW10, kinetochore associate... 32.7 0.30
tgo:TGME49_105790 hypothetical protein 29.3 3.1
dre:414331 cyp51, wu:fb66a09; cytochrome P450, family 51 (EC:1... 28.9 4.5
> tgo:TGME49_031910 ATP synthase gamma chain, putative (EC:3.6.3.14);
K02136 F-type H+-transporting ATPase subunit gamma [EC:3.6.3.14]
Length=314
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 43/66 (65%), Positives = 52/66 (78%), Gaps = 0/66 (0%)
Query 1 EFEPELIDIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFN 60
EFEPE D+WKDLQ F AC GCML+ IA+EQ+ARM+AM+ AS+NA EM+SSL L +N
Sbjct 233 EFEPEKTDVWKDLQDFYYACTVFGCMLDNIASEQSARMSAMDNASTNAGEMISSLTLRYN 292
Query 61 RARQPK 66
RARQ K
Sbjct 293 RARQAK 298
> bbo:BBOV_IV005350 23.m06133; ATP synthase F1 gamma subunit (EC:3.6.3.14);
K02136 F-type H+-transporting ATPase subunit gamma
[EC:3.6.3.14]
Length=310
Score = 89.4 bits (220), Expect = 3e-18, Method: Composition-based stats.
Identities = 40/66 (60%), Positives = 54/66 (81%), Gaps = 0/66 (0%)
Query 1 EFEPELIDIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFN 60
E EPE+ D++ DL +F L CC +GCML+++AAEQ+ARM+AM+ AS+NA++ML SL L FN
Sbjct 230 ETEPEMADLFPDLYEFYLTCCVYGCMLDSLAAEQSARMSAMDNASTNASDMLQSLTLKFN 289
Query 61 RARQPK 66
RARQ K
Sbjct 290 RARQSK 295
> tpv:TP01_0201 ATP synthase F1 subunit gamma; K02136 F-type H+-transporting
ATPase subunit gamma [EC:3.6.3.14]
Length=314
Score = 78.6 bits (192), Expect = 5e-15, Method: Composition-based stats.
Identities = 35/66 (53%), Positives = 49/66 (74%), Gaps = 0/66 (0%)
Query 1 EFEPELIDIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFN 60
E EP++ + D +F L C +GCML+++A+EQ+ARM+AM+ ASSNA +MLS L L +N
Sbjct 234 ETEPDIGKFYPDFYEFYLTSCIYGCMLDSLASEQSARMSAMDNASSNATDMLSKLTLKYN 293
Query 61 RARQPK 66
RARQ K
Sbjct 294 RARQSK 299
> pfa:PF13_0061 ATP synthase gamma chain, mitochondrial precursor,
putative (EC:3.6.3.14); K02136 F-type H+-transporting ATPase
subunit gamma [EC:3.6.3.14]
Length=311
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 33/66 (50%), Positives = 48/66 (72%), Gaps = 0/66 (0%)
Query 1 EFEPELIDIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFN 60
EFEPE+ I+KD+ QF + ++ +A+EQ+ARMTAM+ ASS+A +ML++L L +N
Sbjct 231 EFEPEMDYIFKDIYQFYFTSILYNSIIQNLASEQSARMTAMDNASSSATDMLNALSLRYN 290
Query 61 RARQPK 66
RARQ K
Sbjct 291 RARQSK 296
> xla:380173 atp5c1, MGC53337; ATP synthase, H+ transporting,
mitochondrial F1 complex, gamma polypeptide 1 (EC:3.6.1.14);
K02136 F-type H+-transporting ATPase subunit gamma [EC:3.6.3.14]
Length=294
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 26/57 (45%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 8 DIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFNRARQ 64
D+ ++ Q+F LA + + + +EQ+ARMTAM+ AS NA+EM+ L L+FNR RQ
Sbjct 221 DVLRNYQEFTLANIIYYTLKESTTSEQSARMTAMDNASKNASEMIDKLTLTFNRTRQ 277
> xla:414720 hypothetical protein MGC82276; K02136 F-type H+-transporting
ATPase subunit gamma [EC:3.6.3.14]
Length=294
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 26/57 (45%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 8 DIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFNRARQ 64
D+ ++ Q+F LA + + + +EQ+ARMTAM+ AS NA+EM+ L L+FNR RQ
Sbjct 221 DVLRNYQEFTLANIIYYTLKESTTSEQSARMTAMDNASKNASEMIDKLTLTFNRTRQ 277
> dre:336957 atp5c1, wu:fk34c05, zgc:73209; ATP synthase, H+ transporting,
mitochondrial F1 complex, gamma polypeptide 1 (EC:3.6.1.14);
K02136 F-type H+-transporting ATPase subunit gamma
[EC:3.6.3.14]
Length=292
Score = 57.8 bits (138), Expect = 9e-09, Method: Composition-based stats.
Identities = 26/57 (45%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 8 DIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFNRARQ 64
D+ ++ Q+FAL + + + +EQ+ARMTAM+ AS NA+EM+ L L+FNR RQ
Sbjct 219 DVLRNYQEFALVNIIYFGLKESTTSEQSARMTAMDSASKNASEMIDKLTLTFNRTRQ 275
> hsa:509 ATP5C1, ATP5C, ATP5CL1; ATP synthase, H+ transporting,
mitochondrial F1 complex, gamma polypeptide 1 (EC:3.6.1.14);
K02136 F-type H+-transporting ATPase subunit gamma [EC:3.6.3.14]
Length=298
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 25/57 (43%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 8 DIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFNRARQ 64
D+ ++ Q++ LA + + + +EQ+ARMTAM+ AS NA+EM+ L L+FNR RQ
Sbjct 224 DVLQNYQEYNLANIIYYSLKESTTSEQSARMTAMDNASKNASEMIDKLTLTFNRTRQ 280
> mmu:11949 Atp5c1, 1700094F02Rik; ATP synthase, H+ transporting,
mitochondrial F1 complex, gamma polypeptide 1 (EC:3.6.1.14);
K02136 F-type H+-transporting ATPase subunit gamma [EC:3.6.3.14]
Length=274
Score = 54.3 bits (129), Expect = 8e-08, Method: Composition-based stats.
Identities = 24/57 (42%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 8 DIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFNRARQ 64
D+ ++ Q++ LA + + + +EQ+ARMTAM+ AS NA++M+ L L+FNR RQ
Sbjct 200 DVLQNYQEYNLANLIYYSLKESTTSEQSARMTAMDNASKNASDMIDKLTLTFNRTRQ 256
> ath:AT2G33040 ATP synthase gamma chain, mitochondrial (ATPC);
K02136 F-type H+-transporting ATPase subunit gamma [EC:3.6.3.14]
Length=325
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 23/57 (40%), Positives = 35/57 (61%), Gaps = 0/57 (0%)
Query 8 DIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFNRARQ 64
+I ++L +F +C +L +E ARM+AM+ +S NA EML L L++NR RQ
Sbjct 248 EILQNLAEFQFSCVMFNAVLENACSEMGARMSAMDSSSRNAGEMLDRLTLTYNRTRQ 304
> eco:b3733 atpG, ECK3726, JW3711, papC, uncG; F1 sector of membrane-bound
ATP synthase, gamma subunit (EC:3.6.3.14); K02115
F-type H+-transporting ATPase subunit gamma [EC:3.6.3.14]
Length=287
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 43/71 (60%), Gaps = 6/71 (8%)
Query 2 FEPE---LIDIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLS 58
+EP+ L+D L+++ + + G + N +A+EQAARM AM+ A+ N ++ L+L
Sbjct 208 YEPDPKALLDTL--LRRYVESQVYQGVVEN-LASEQAARMVAMKAATDNGGSLIKELQLV 264
Query 59 FNRARQPKSLQ 69
+N+ARQ Q
Sbjct 265 YNKARQASITQ 275
> ath:AT4G04640 ATPC1; ATPC1; enzyme regulator; K02115 F-type
H+-transporting ATPase subunit gamma [EC:3.6.3.14]
Length=373
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 24/66 (36%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 1 EFEPELIDIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFN 60
+FE + + I L L + ++A+E AARM+AM AS NA+++ SL + +N
Sbjct 292 QFEQDPVQILDALLPLYLNSQILRALQESLASELAARMSAMSSASDNASDLKKSLSMVYN 351
Query 61 RARQPK 66
R RQ K
Sbjct 352 RKRQAK 357
> ath:AT1G15700 ATPC2; ATPC2; enzyme regulator; K02115 F-type
H+-transporting ATPase subunit gamma [EC:3.6.3.14]
Length=386
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 1 EFEPELIDIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFN 60
+FE + + I + L + ++A+E A+RM AM A+ NA E+ +L +++N
Sbjct 303 QFEQDPVQILDAMMPLYLNSQILRALQESLASELASRMNAMSNATDNAVELKKNLTMAYN 362
Query 61 RARQPK 66
RARQ K
Sbjct 363 RARQAK 368
> sce:YBR039W ATP3; Atp3p (EC:3.6.3.14); K02136 F-type H+-transporting
ATPase subunit gamma [EC:3.6.3.14]
Length=311
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 8 DIWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSLRLSFNRARQ 64
++ +DL ++ LA M AAE +AR AM+ AS NA +M++ + +NR RQ
Sbjct 237 NVPRDLFEYTLANQMLTAMAQGYAAEISARRNAMDNASKNAGDMINRYSILYNRTRQ 293
> mmu:26951 Zw10, 6330566F14Rik, MmZw10; ZW10 homolog (Drosophila),
centromere/kinetochore protein; K11578 centromere/kinetochore
protein ZW10
Length=779
Score = 35.8 bits (81), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 9 IWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSL 55
+W+D+ + C G +LN AE +R+TA+E S+ + L SL
Sbjct 639 VWQDVLPVNIYCKAMGTLLNTAIAEMMSRITALEDISTEDGDRLYSL 685
> xla:443575 zw10, MGC181630; centromere/kinetochore protein zw10;
K11578 centromere/kinetochore protein ZW10
Length=776
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 9 IWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSL 55
+W+D+ A+ C G +LN E +++TA+E S+ E L SL
Sbjct 636 VWQDVLPVAIYCKAMGTLLNTAIVEMISKITALEDISTEDGERLYSL 682
> hsa:9183 ZW10, HZW10, KNTC1AP, MGC149821; ZW10, kinetochore
associated, homolog (Drosophila); K11578 centromere/kinetochore
protein ZW10
Length=779
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 9 IWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSL 55
+W+D+ + C G +LN +E ++TA+E S+ + L SL
Sbjct 639 VWQDVLPVNIYCKAMGTLLNTAISEVIGKITALEDISTEDGDRLYSL 685
> xla:445850 zw10, MGC114854, xzw10; ZW10, kinetochore associated,
homolog; K11578 centromere/kinetochore protein ZW10
Length=776
Score = 32.7 bits (73), Expect = 0.30, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 9 IWKDLQQFALACCFHGCMLNAIAAEQAARMTAMEGASSNAAEMLSSL 55
+W+D+ A+ C G +LN E ++TA+E S+ E L +L
Sbjct 636 VWQDVLPVAIYCKAMGTLLNTTIVEMIGKITALEDISTEDGERLYTL 682
> tgo:TGME49_105790 hypothetical protein
Length=1918
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 10/25 (40%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 12 DLQQFALACCFHGCMLNAIAAEQAA 36
D+QQ + ++GC+LN ++ +QAA
Sbjct 1843 DMQQLMMYAQYYGCLLNGMSEQQAA 1867
> dre:414331 cyp51, wu:fb66a09; cytochrome P450, family 51 (EC:1.14.14.1);
K05917 cytochrome P450, family 51 (sterol 14-demethylase)
[EC:1.14.13.70]
Length=499
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 15/51 (29%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query 3 EPELIDIWKDLQQFALACCFHGCMLNAIAAEQAARMTA-MEGASSNAAEML 52
E L D +L + C HGC + ++ E+ A++ A ++G ++AA +L
Sbjct 183 ERNLFDALSELIILTASRCLHGCEIRSLLDERVAQLYADLDGGFTHAAWLL 233
Lambda K H
0.322 0.128 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2029389012
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40