bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5329_orf1
Length=105
Score E
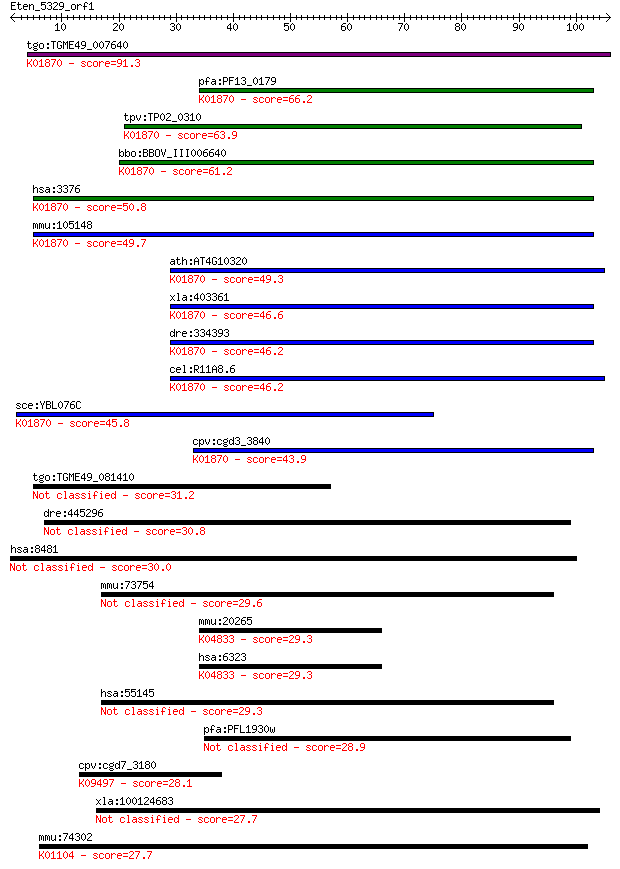
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_007640 isoleucine-tRNA synthetase, putative (EC:6.1... 91.3 7e-19
pfa:PF13_0179 isoleucine-tRNA ligase, putative (EC:6.1.1.5); K... 66.2 3e-11
tpv:TP02_0310 isoleucyl-tRNA synthetase; K01870 isoleucyl-tRNA... 63.9 1e-10
bbo:BBOV_III006640 17.m07588; isoleucyl-tRNA synthetase family... 61.2 7e-10
hsa:3376 IARS, FLJ20736, IARS1, ILERS, ILRS, IRS, PRO0785; iso... 50.8 1e-06
mmu:105148 Iars, 2510016L12Rik, AI327140, AU044614, E430001P04... 49.7 2e-06
ath:AT4G10320 isoleucyl-tRNA synthetase, putative / isoleucine... 49.3 3e-06
xla:403361 iars, MGC68929; isoleucyl-tRNA synthetase; K01870 i... 46.6 2e-05
dre:334393 iars, fi46h05, wu:fi46h05, zgc:63790; isoleucyl-tRN... 46.2 2e-05
cel:R11A8.6 irs-1; Isoleucyl tRNA Synthetase family member (ir... 46.2 2e-05
sce:YBL076C ILS1; Ils1p (EC:6.1.1.5); K01870 isoleucyl-tRNA sy... 45.8 4e-05
cpv:cgd3_3840 isoleucine-tRNA synthetase ; K01870 isoleucyl-tR... 43.9 1e-04
tgo:TGME49_081410 pyrophosphate dependent phosphofructokinase,... 31.2 0.73
dre:445296 merit40, nba1; zgc:100909 30.8 1.1
hsa:8481 OFD1, 71-7A, CXorf5, JBTS10, MGC117039, MGC117040, SG... 30.0 1.6
mmu:73754 Thap1, 4833431A01Rik, AW490810; THAP domain containi... 29.6 2.3
mmu:20265 Scn1a, B230332M13, Nav1.1; sodium channel, voltage-g... 29.3 2.8
hsa:6323 SCN1A, EIEE6, FEB3, FEB3A, FHM3, GEFSP2, HBSCI, NAC1,... 29.3 2.8
hsa:55145 THAP1, DYT6, FLJ10477, MGC33014; THAP domain contain... 29.3 3.4
pfa:PFL1930w conserved Plasmodium protein 28.9 4.6
cpv:cgd7_3180 T complex chaperonin ; K09497 T-complex protein ... 28.1 7.7
xla:100124683 tax1bp1, MGC80176; Tax1 binding protein 1 27.7
mmu:74302 Mtmr3, 1700092A20Rik, AI255150, AW557713, FYVE-DSP1,... 27.7 8.9
> tgo:TGME49_007640 isoleucine-tRNA synthetase, putative (EC:6.1.1.5);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1103
Score = 91.3 bits (225), Expect = 7e-19, Method: Composition-based stats.
Identities = 50/104 (48%), Positives = 68/104 (65%), Gaps = 2/104 (1%)
Query 4 LLTPQEATLQRQLP--QSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLR 61
+L + TL+R L +PNL D + V+ MDFT D SLQRKA+ARE+ANRVQKLR
Sbjct 965 VLGADDLTLRRTLNTGDKADPNLVVEGDNSVVVLMDFTLDDSLQRKALAREVANRVQKLR 1024
Query 62 KQLQLQQQDEVLVFAAADDAALKDILIQEKEYIESCLRRPLLLQ 105
KQ L Q D+V + A ++D+ + +L +E YI +CLRR L L+
Sbjct 1025 KQHNLSQTDDVKMHAFSEDSEFQAMLQEESAYICACLRRGLHLE 1068
> pfa:PF13_0179 isoleucine-tRNA ligase, putative (EC:6.1.1.5);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1272
Score = 66.2 bits (160), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/69 (47%), Positives = 41/69 (59%), Gaps = 0/69 (0%)
Query 34 IAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFAAADDAALKDILIQEKEY 93
I MDFT D L+ A AREI N +QK+RK L L Q VL+ D K+ ++ E EY
Sbjct 1169 ILMDFTADQQLENMASAREICNHIQKIRKNLSLNQNSPVLMHIYIYDQTFKNHMLNENEY 1228
Query 94 IESCLRRPL 102
I+ CLRR L
Sbjct 1229 IKKCLRRNL 1237
> tpv:TP02_0310 isoleucyl-tRNA synthetase; K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1129
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 29/80 (36%), Positives = 49/80 (61%), Gaps = 0/80 (0%)
Query 21 NPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFAAADD 80
N N D N + ++ T+D +L+ KA+ RE+ANRVQK+RK+L L D++ V+ DD
Sbjct 1017 NNNFDAESDNNLTVILELTKDDTLEYKALTREVANRVQKMRKELGLSIDDDITVYIYTDD 1076
Query 81 AALKDILIQEKEYIESCLRR 100
L ++L + E ++ L++
Sbjct 1077 LELFNMLESQTESLKKILKK 1096
> bbo:BBOV_III006640 17.m07588; isoleucyl-tRNA synthetase family
protein (EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1080
Score = 61.2 bits (147), Expect = 7e-10, Method: Composition-based stats.
Identities = 29/83 (34%), Positives = 46/83 (55%), Gaps = 0/83 (0%)
Query 20 TNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFAAAD 79
+NPN+ + ++ + +DFT D SLQ A RE+ NRVQKLRK+L+L D+V ++
Sbjct 967 SNPNVDGDSNREVAVLLDFTSDESLQWMAYGREVCNRVQKLRKELRLSVNDKVNIYVQLA 1026
Query 80 DAALKDILIQEKEYIESCLRRPL 102
D + +Y+ LR +
Sbjct 1027 SDECFDKFSAQSDYLSKTLRHAI 1049
> hsa:3376 IARS, FLJ20736, IARS1, ILERS, ILRS, IRS, PRO0785; isoleucyl-tRNA
synthetase (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1262
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 30/102 (29%), Positives = 53/102 (51%), Gaps = 4/102 (3%)
Query 5 LTPQEATLQRQLPQSTNPNLGF--NCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRK 62
L ++ L Q+T F + D ++ +D T D S+ + +ARE+ NR+QKLRK
Sbjct 940 LHDEDIRLMYTFDQATGGTAQFEAHSDAQALVLLDVTPDQSMVDEGMAREVINRIQKLRK 999
Query 63 QLQLQQQDEVLVF--AAADDAALKDILIQEKEYIESCLRRPL 102
+ L DE+ V+ A ++ L ++ E+I + ++ PL
Sbjct 1000 KCNLVPTDEITVYYKAKSEGTYLNSVIESHTEFIFTTIKAPL 1041
> mmu:105148 Iars, 2510016L12Rik, AI327140, AU044614, E430001P04Rik,
ILRS; isoleucine-tRNA synthetase (EC:6.1.1.5); K01870
isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1262
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 29/102 (28%), Positives = 54/102 (52%), Gaps = 4/102 (3%)
Query 5 LTPQEATLQRQLPQSTNPNLGF--NCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRK 62
L ++ L Q+T F + D ++ +D T D S+ + +ARE+ NR+QKLRK
Sbjct 940 LHEEDIRLMYTFDQATGGTAQFEAHSDAQALVLLDVTPDQSMVDEGMAREVINRIQKLRK 999
Query 63 QLQLQQQDEVLVF--AAADDAALKDILIQEKEYIESCLRRPL 102
+ L DE+ V+ A ++ L +++ ++I + ++ PL
Sbjct 1000 KCNLVPTDEITVYYNAKSEGRYLNNVIESHTDFIFATIKAPL 1041
> ath:AT4G10320 isoleucyl-tRNA synthetase, putative / isoleucine--tRNA
ligase, putative (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1190
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 3/79 (3%)
Query 29 DKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFAAA---DDAALKD 85
D + ++ +D D SL AREI NR+QKLRK+ L+ D V V+ + D++ K
Sbjct 992 DGDVLVILDLRADDSLVEAGFAREIVNRIQKLRKKSGLEPTDFVEVYFQSLDEDESVSKQ 1051
Query 86 ILIQEKEYIESCLRRPLLL 104
+L+ +++ I+ + LLL
Sbjct 1052 VLVSQEQNIKDSIGSTLLL 1070
> xla:403361 iars, MGC68929; isoleucyl-tRNA synthetase; K01870
isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1259
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 45/76 (59%), Gaps = 2/76 (2%)
Query 29 DKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVF--AAADDAALKDI 86
D ++ +D T D S+ + +ARE+ NR+QKLRK+ L DE+ ++ + + L+ +
Sbjct 966 DAQVLVLLDVTPDQSMVDEGVAREVINRIQKLRKKGNLVPTDEITIYYQSLPEGNYLQSV 1025
Query 87 LIQEKEYIESCLRRPL 102
+ + ++I + ++ PL
Sbjct 1026 IERHTDFILATVKSPL 1041
> dre:334393 iars, fi46h05, wu:fi46h05, zgc:63790; isoleucyl-tRNA
synthetase (EC:6.1.1.5); K01870 isoleucyl-tRNA synthetase
[EC:6.1.1.5]
Length=1271
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 41/76 (53%), Gaps = 2/76 (2%)
Query 29 DKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFAAADDAA--LKDI 86
D ++ +D T D ++ + +ARE+ NR+QKLRK+ L DE+ VF A L +
Sbjct 966 DAQVLVLLDVTPDQAMLDEGVAREVINRIQKLRKKGHLVPSDEITVFYRCQPAGEYLHQV 1025
Query 87 LIQEKEYIESCLRRPL 102
+ ++I + + PL
Sbjct 1026 IEAHTDFILATTKAPL 1041
> cel:R11A8.6 irs-1; Isoleucyl tRNA Synthetase family member (irs-1);
K01870 isoleucyl-tRNA synthetase [EC:6.1.1.5]
Length=1141
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/78 (33%), Positives = 41/78 (52%), Gaps = 2/78 (2%)
Query 29 DKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQD--EVLVFAAADDAALKDI 86
D ++ +D TED SL + + RE+ NRVQ+LRKQ +L D V + + D+ L +
Sbjct 958 DAKTIVMIDTTEDESLIEEGLCREVTNRVQRLRKQAKLVSTDTAHVHIVVSPADSQLAKV 1017
Query 87 LIQEKEYIESCLRRPLLL 104
+ + I S P+ L
Sbjct 1018 VAAKLNDIVSATGTPIKL 1035
> sce:YBL076C ILS1; Ils1p (EC:6.1.1.5); K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1072
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 40/74 (54%), Gaps = 1/74 (1%)
Query 2 GILLTPQEATLQRQLPQST-NPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKL 60
GI L + R LP+S D++ +I MD L+ + +ARE+ NR+QKL
Sbjct 939 GIELVKGDLNAIRGLPESAVQAGQETRTDQDVLIIMDTNIYSELKSEGLARELVNRIQKL 998
Query 61 RKQLQLQQQDEVLV 74
RK+ L+ D+VLV
Sbjct 999 RKKCGLEATDDVLV 1012
> cpv:cgd3_3840 isoleucine-tRNA synthetase ; K01870 isoleucyl-tRNA
synthetase [EC:6.1.1.5]
Length=1100
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 39/75 (52%), Gaps = 5/75 (6%)
Query 33 VIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFA-----AADDAALKDIL 87
VI +DFT D + A +RE+AN++QK+RK+ + V ++ + ++
Sbjct 989 VIGLDFTSDIEFENMAYSRELANKIQKIRKEKNMDPDANVTIYLQLISNKNESKKFHTVM 1048
Query 88 IQEKEYIESCLRRPL 102
+Y++ +R+P+
Sbjct 1049 TSYNDYLQKIIRKPI 1063
> tgo:TGME49_081410 pyrophosphate dependent phosphofructokinase,
putative (EC:2.7.1.90)
Length=1720
Score = 31.2 bits (69), Expect = 0.73, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 5 LTPQEATLQRQLPQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANR 56
LTP A L LP++ L FN D+ + + + L ++ + RE+A R
Sbjct 1167 LTPWSAALFSSLPKAIQWQLAFNKDQTYRVDLTAIDTEVLLKRLVERELARR 1218
> dre:445296 merit40, nba1; zgc:100909
Length=396
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 25/104 (24%), Positives = 48/104 (46%), Gaps = 12/104 (11%)
Query 7 PQEATLQRQLPQSTNPNLG---FNCDKNCVIAMDFTEDPSLQR-------KAIAREIANR 56
P + T+ Q+P S +L NC + +I +D +E+ SLQ+ K A I+ +
Sbjct 109 PSQPTMPTQIPPSAELHLRAPRVNCPEKVIICLDLSEEMSLQKLESINGSKTNALNISQK 168
Query 57 VQKL--RKQLQLQQQDEVLVFAAADDAALKDILIQEKEYIESCL 98
+ ++ R + ++ ++ E + DDA + + SCL
Sbjct 169 MIEMFVRTKHKIDKRHEFALVVVNDDAMWLSGFTSDPRELCSCL 212
> hsa:8481 OFD1, 71-7A, CXorf5, JBTS10, MGC117039, MGC117040,
SGBS2; oral-facial-digital syndrome 1
Length=1012
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Query 1 NGILLTPQEATLQRQLPQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKL 60
N + P+++ + R + N N+ C N I+ DF +P Q +AR +A+R+
Sbjct 555 NEVYCNPKQSVIDRSVNGLINGNV-VPC--NGEISGDFLNNPFKQENVLARMVASRITNY 611
Query 61 -RKQLQLQQQDEVLVFAAADDAALKDILIQEKEYIESCLR 99
++ D L F A A +K+ L QE E +E R
Sbjct 612 PTAWVEGSSPDSDLEFVANTKARVKE-LQQEAERLEKAFR 650
> mmu:73754 Thap1, 4833431A01Rik, AW490810; THAP domain containing,
apoptosis associated protein 1
Length=210
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 29/92 (31%), Positives = 43/92 (46%), Gaps = 19/92 (20%)
Query 17 PQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQ----------- 65
P T NL CD N + ED QRK I ++ +V+KLRK+L+
Sbjct 119 PLQTPDNLSVFCDHNYTV-----EDTMHQRKRIL-QLEQQVEKLRKKLKTAQQRCRRQER 172
Query 66 -LQQQDEVLVFA-AADDAALKDILIQEKEYIE 95
L++ EV+ F DDA+ + +I +Y E
Sbjct 173 QLEKLKEVVHFQREKDDASERGYVILPNDYFE 204
> mmu:20265 Scn1a, B230332M13, Nav1.1; sodium channel, voltage-gated,
type I, alpha; K04833 voltage-gated sodium channel type
I alpha
Length=1998
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 2/34 (5%)
Query 34 IAMDFTEDPSLQRKA--IAREIANRVQKLRKQLQ 65
++MDF EDPS +++A IA + N V++L + Q
Sbjct 688 VSMDFLEDPSQRQRAMSIASILTNTVEELEESRQ 721
> hsa:6323 SCN1A, EIEE6, FEB3, FEB3A, FHM3, GEFSP2, HBSCI, NAC1,
Nav1.1, SCN1, SMEI; sodium channel, voltage-gated, type I,
alpha subunit; K04833 voltage-gated sodium channel type I
alpha
Length=2009
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 2/34 (5%)
Query 34 IAMDFTEDPSLQRKA--IAREIANRVQKLRKQLQ 65
++MDF EDPS +++A IA + N V++L + Q
Sbjct 699 VSMDFLEDPSQRQRAMSIASILTNTVEELEESRQ 732
> hsa:55145 THAP1, DYT6, FLJ10477, MGC33014; THAP domain containing,
apoptosis associated protein 1
Length=213
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 42/92 (45%), Gaps = 19/92 (20%)
Query 17 PQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQ----------- 65
P T NL CD N + ED QRK I ++ +V+KLRK+L+
Sbjct 122 PLQTPVNLSVFCDHNYTV-----EDTMHQRKRI-HQLEQQVEKLRKKLKTAQQRCRRQER 175
Query 66 -LQQQDEVLVF-AAADDAALKDILIQEKEYIE 95
L++ EV+ F DD + + +I +Y E
Sbjct 176 QLEKLKEVVHFQKEKDDVSERGYVILPNDYFE 207
> pfa:PFL1930w conserved Plasmodium protein
Length=5767
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 14/64 (21%), Positives = 38/64 (59%), Gaps = 1/64 (1%)
Query 35 AMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVFAAADDAALKDILIQEKEYI 94
++D ++ +++ + E+ NR+ ++ +L+++ + L++ ++ +KDILI+ E +
Sbjct 1466 SLDTIDELNMKYEEEKNEMLNRINEVENKLKIKMNENNLLYEKYENN-MKDILIERDERV 1524
Query 95 ESCL 98
CL
Sbjct 1525 SDCL 1528
> cpv:cgd7_3180 T complex chaperonin ; K09497 T-complex protein
1 subunit epsilon
Length=556
Score = 28.1 bits (61), Expect = 7.7, Method: Composition-based stats.
Identities = 12/25 (48%), Positives = 18/25 (72%), Gaps = 1/25 (4%)
Query 13 QRQLPQSTNPNLGFNCDKNCVIAMD 37
Q+Q+ Q +PN G +CDK CV +M+
Sbjct 495 QKQVSQD-DPNYGIDCDKLCVESME 518
> xla:100124683 tax1bp1, MGC80176; Tax1 binding protein 1
Length=840
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 42/88 (47%), Gaps = 6/88 (6%)
Query 16 LPQSTNPNLGFNCDKNCVIAMDFTEDPSLQRKAIAREIANRVQKLRKQLQLQQQDEVLVF 75
+ Q T P F D V D E Q + ++ EIA + +K RK Q+ +++
Sbjct 509 MNQCTPPVSPFTSD--TVPEFDIRE----QVREMSLEIAEKTEKYRKCKQMLAEEKARCS 562
Query 76 AAADDAALKDILIQEKEYIESCLRRPLL 103
A ADD A ++ +E+ + ++R L+
Sbjct 563 AYADDVAKMELKWKEQLKVNDSIKRQLI 590
> mmu:74302 Mtmr3, 1700092A20Rik, AI255150, AW557713, FYVE-DSP1,
MGC78036, ZFYVE10, mKIAA0371; myotubularin related protein
3 (EC:3.1.3.48); K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=1159
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 30/109 (27%), Positives = 43/109 (39%), Gaps = 14/109 (12%)
Query 6 TPQEATLQRQLPQSTNPNLGFNCDKNCVIAMDFTEDPSLQRK-------------AIARE 52
+P E L R + NL C+ +A + DPSL K A A E
Sbjct 599 SPDEPPLSRLPKTRSFDNLTTTCENMVPLASRRSSDPSLNEKWQEHGRSLELSSFASAGE 658
Query 53 IANRVQKLRKQLQLQQQDEVLVFAAADDAALKDILIQEKEYIESCLRRP 101
+ LRK +L E+ V A + +++IL QE ES + P
Sbjct 659 EVPAMDSLRKPSRLLGGAELSVAAGVAEGQMENIL-QEATKEESGVEEP 706
Lambda K H
0.320 0.134 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2017521068
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40