bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5240_orf1
Length=109
Score E
Sequences producing significant alignments: (Bits) Value
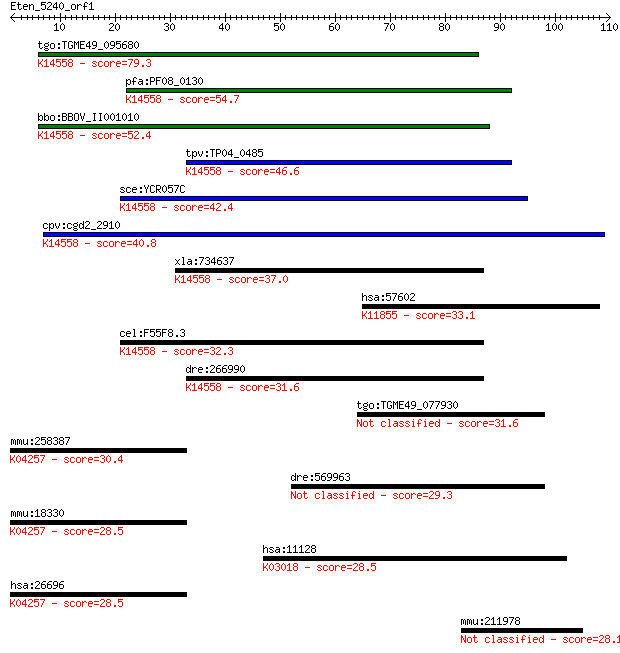
tgo:TGME49_095680 periodic tryptophan protein PWP2, putative ;... 79.3 3e-15
pfa:PF08_0130 WD-repeat protein, putative; K14558 periodic try... 54.7 8e-08
bbo:BBOV_II001010 18.m06073; periodic tryptophan protein 2-lik... 52.4 4e-07
tpv:TP04_0485 hypothetical protein; K14558 periodic tryptophan... 46.6 2e-05
sce:YCR057C PWP2, UTP1, YCR055C, YCR058C; Conserved 90S pre-ri... 42.4 4e-04
cpv:cgd2_2910 hypothetical protein ; K14558 periodic tryptopha... 40.8 0.001
xla:734637 hypothetical protein MGC115367; K14558 periodic try... 37.0 0.015
hsa:57602 USP36, DUB1; ubiquitin specific peptidase 36 (EC:3.4... 33.1 0.24
cel:F55F8.3 hypothetical protein; K14558 periodic tryptophan p... 32.3 0.39
dre:266990 pwp2h, cb471, zgc:56063; PWP2 periodic tryptophan p... 31.6 0.60
tgo:TGME49_077930 hypothetical protein 31.6 0.73
mmu:258387 Olfr720, MOR274-2; olfactory receptor 720; K04257 o... 30.4 1.5
dre:569963 syntaphilin-like 29.3 3.4
mmu:18330 Olfr31, MGC123774, MGC123790, MOR274-1, MTPCR53; olf... 28.5 5.0
hsa:11128 POLR3A, RPC1, RPC155, hRPC155; polymerase (RNA) III ... 28.5 5.1
hsa:26696 OR2T1, OR1-25; olfactory receptor, family 2, subfami... 28.5 5.1
mmu:211978 Zfyve26, 4930465A13, 9330197E15Rik, A630028O16Rik, ... 28.1 7.0
> tgo:TGME49_095680 periodic tryptophan protein PWP2, putative
; K14558 periodic tryptophan protein 2
Length=1266
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 40/82 (48%), Positives = 53/82 (64%), Gaps = 2/82 (2%)
Query 6 FFIYGIDLFGGILGNYGVTE--TGMSLPFLTKNVTTSNIAKALHNEEYAKAFLLALVANN 63
+I+ +D GI +G + P LT NVTT +I++AL EYAKAF+L+LV N+
Sbjct 1044 LYIFSLDSKSGIYAAFGTSSRLPCAPPPMLTANVTTGSISRALERREYAKAFILSLVLND 1103
Query 64 RPTLLCVYERIPAASVPLVCSS 85
PTLL +YE IP +SV LVCSS
Sbjct 1104 LPTLLSIYEAIPPSSVALVCSS 1125
> pfa:PF08_0130 WD-repeat protein, putative; K14558 periodic tryptophan
protein 2
Length=1121
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 28/71 (39%), Positives = 41/71 (57%), Gaps = 1/71 (1%)
Query 22 GVTETGMSLP-FLTKNVTTSNIAKALHNEEYAKAFLLALVANNRPTLLCVYERIPAASVP 80
G+ + +P FLT+NV +N+ +L +EY KAF+L+L NN +L VYE IP +P
Sbjct 936 GLISPNIYVPKFLTQNVNLNNLKNSLKKKEYMKAFILSLALNNYENILEVYENIPYNIIP 995
Query 81 LVCSSLSAALL 91
L L+ L
Sbjct 996 LCVKVLTKPFL 1006
> bbo:BBOV_II001010 18.m06073; periodic tryptophan protein 2-like
protein; K14558 periodic tryptophan protein 2
Length=988
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 32/89 (35%), Positives = 51/89 (57%), Gaps = 11/89 (12%)
Query 6 FFIYGIDLF-------GGILGNYGVTETGMSLPFLTKNVTTSNIAKALHNEEYAKAFLLA 58
++Y +D F G + + G+ + + LTKNVT N+ AL + ++ KAF+LA
Sbjct 799 LYVYSMDAFIKTPNYLGEVFKSVGLFQPQL----LTKNVTVGNVLGALESGDHIKAFILA 854
Query 59 LVANNRPTLLCVYERIPAASVPLVCSSLS 87
L N+ TLL YE +PA+SV V +S++
Sbjct 855 LALNDFNTLLRCYESVPASSVSHVVASIA 883
> tpv:TP04_0485 hypothetical protein; K14558 periodic tryptophan
protein 2
Length=968
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 23/59 (38%), Positives = 34/59 (57%), Gaps = 0/59 (0%)
Query 33 LTKNVTTSNIAKALHNEEYAKAFLLALVANNRPTLLCVYERIPAASVPLVCSSLSAALL 91
+TK+VT S+I L ++ AF+LAL NN +L YE +P + V SSL + L+
Sbjct 818 ITKSVTKSSILSHLEKNDFVGAFILALAMNNFKIMLKAYEAVPTGKIASVVSSLDSTLV 876
> sce:YCR057C PWP2, UTP1, YCR055C, YCR058C; Conserved 90S pre-ribosomal
component essential for proper endonucleolytic cleavage
of the 35 S rRNA precursor at A0, A1, and A2 sites; contains
eight WD-repeats; PWP2 deletion leads to defects in cell
cycle and bud morphogenesis; K14558 periodic tryptophan
protein 2
Length=923
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 39/75 (52%), Gaps = 1/75 (1%)
Query 21 YGVTETGMSLPF-LTKNVTTSNIAKALHNEEYAKAFLLALVANNRPTLLCVYERIPAASV 79
Y +T + PF L +VT + +AL +++ A ++A N + VYE IP +
Sbjct 704 YSTNDTILFDPFDLDVDVTPHSTVEALREKQFLNALVMAFRLNEEYLINKVYEAIPIKEI 763
Query 80 PLVCSSLSAALLPSL 94
PLV S++ A LP +
Sbjct 764 PLVASNIPAIYLPRI 778
> cpv:cgd2_2910 hypothetical protein ; K14558 periodic tryptophan
protein 2
Length=1003
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 28/108 (25%), Positives = 51/108 (47%), Gaps = 9/108 (8%)
Query 7 FIYGIDLFGGILGNYGVTETGMSL------PFLTKNVTTSNIAKALHNEEYAKAFLLALV 60
F++ ID G +Y + T +++ +TK V SNI L ++ + +LAL
Sbjct 790 FLFCIDTLGN---SYSGSSTHLNMLDTFKKQIMTKEVNVSNIDHFLKEGDFLRGLILALA 846
Query 61 ANNRPTLLCVYERIPAASVPLVCSSLSAALLPSLSSPPQCVTVKKAQQ 108
NN L Y +IP S+ + ++ + LP++ + + VT +Q
Sbjct 847 MNNFNILAKAYVQIPITSIDYIVQNIVSFFLPNILNFLRIVTSPNNKQ 894
> xla:734637 hypothetical protein MGC115367; K14558 periodic tryptophan
protein 2
Length=895
Score = 37.0 bits (84), Expect = 0.015, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Query 31 PF-LTKNVTTSNIAKALHNEEYAKAFLLALVANNRPTLLCVYERIPAASVPLVCSSL 86
PF L + VT + +A+ E+ +A ++A+ N L E +P A + ++CSSL
Sbjct 730 PFDLDEEVTAGGVRRAVRGGEWTRAIVMAMRLNEESLLRETLESVPCADIAVLCSSL 786
> hsa:57602 USP36, DUB1; ubiquitin specific peptidase 36 (EC:3.4.19.12);
K11855 ubiquitin carboxyl-terminal hydrolase 36/42
[EC:3.1.2.15]
Length=1123
Score = 33.1 bits (74), Expect = 0.24, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 25/56 (44%), Gaps = 13/56 (23%)
Query 65 PTLLCVYERIPAASVPLVCSSLSAAL-------------LPSLSSPPQCVTVKKAQ 107
PTLL + P S P CSS+S AL LP S PPQ + K+ +
Sbjct 759 PTLLSSTPKPPGTSEPRSCSSISTALPQVNEDLVSLPHQLPEASEPPQSPSEKRKK 814
> cel:F55F8.3 hypothetical protein; K14558 periodic tryptophan
protein 2
Length=910
Score = 32.3 bits (72), Expect = 0.39, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query 21 YGVTETGMSLPFLTKNVTTSNIAK-ALHNEEYAKAFLLALVANNRPTLLCVYERIPAASV 79
Y + M PF + T + + K A+ N +Y+ A + +L NN + E + +
Sbjct 716 YSLDTISMFDPFQLDSQTNAEVIKRAVWNNDYSTAIMASLRLNNAQYITECLESTSISQI 775
Query 80 PLVCSSL 86
PLV SSL
Sbjct 776 PLVASSL 782
> dre:266990 pwp2h, cb471, zgc:56063; PWP2 periodic tryptophan
protein homolog (yeast); K14558 periodic tryptophan protein
2
Length=937
Score = 31.6 bits (70), Expect = 0.60, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 33 LTKNVTTSNIAKALHNEEYAKAFLLALVANNRPTLLCVYERIPAASVPLVCSSL 86
L +VT ++I + + +E+A A LL N + V E IP + +VCSSL
Sbjct 753 LDIDVTPTSIRQQITKKEWASAILLFFRLNEILLIREVLEAIPFDQITVVCSSL 806
> tgo:TGME49_077930 hypothetical protein
Length=1573
Score = 31.6 bits (70), Expect = 0.73, Method: Composition-based stats.
Identities = 17/34 (50%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Query 64 RPTLLCVYERIPAASVPLVCSSLSAALLPSLSSP 97
RPT L R P AS PL C +++++LL SL+SP
Sbjct 433 RPTDLDALIRSPPAS-PLCCIAVASSLLRSLASP 465
> mmu:258387 Olfr720, MOR274-2; olfactory receptor 720; K04257
olfactory receptor
Length=316
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 19/32 (59%), Gaps = 1/32 (3%)
Query 1 RQFCWFFIYGIDLFGGILGNYGVTETGMSLPF 32
R+ CWF I FGG L ++ +T MSLPF
Sbjct 137 RRVCWF-ILASSWFGGALDSFLLTPITMSLPF 167
> dre:569963 syntaphilin-like
Length=626
Score = 29.3 bits (64), Expect = 3.4, Method: Composition-based stats.
Identities = 22/61 (36%), Positives = 29/61 (47%), Gaps = 15/61 (24%)
Query 52 AKAFLL---ALVANNRPTLLCVYERIPAAS------------VPLVCSSLSAALLPSLSS 96
+KA LL A ++ +LL Y R+P +S VPL CSS+ A SLS
Sbjct 254 SKADLLLEAAFLSQETASLLNAYSRLPHSSTYEGLCSSEPRVVPLRCSSIGAGASVSLSH 313
Query 97 P 97
P
Sbjct 314 P 314
> mmu:18330 Olfr31, MGC123774, MGC123790, MOR274-1, MTPCR53; olfactory
receptor 31; K04257 olfactory receptor
Length=317
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 17/32 (53%), Gaps = 1/32 (3%)
Query 1 RQFCWFFIYGIDLFGGILGNYGVTETGMSLPF 32
R+ CW I G FGG L + +T MS PF
Sbjct 137 RRICWIIIAG-SWFGGSLDGFLLTPITMSFPF 167
> hsa:11128 POLR3A, RPC1, RPC155, hRPC155; polymerase (RNA) III
(DNA directed) polypeptide A, 155kDa (EC:2.7.7.6); K03018
DNA-directed RNA polymerase III subunit RPC1 [EC:2.7.7.6]
Length=1390
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 5/60 (8%)
Query 47 HNEEYAKAFLLALVANNRPTLLCVYERIPAASVPLVCSSLSAA-----LLPSLSSPPQCV 101
HN+E A N +L +++RIPA VPL+ + A +L L PP C+
Sbjct 203 HNKEVEPLLGRAQENLNPLVVLNLFKRIPAEDVPLLLMNPEAGKPSDLILTRLLVPPLCI 262
> hsa:26696 OR2T1, OR1-25; olfactory receptor, family 2, subfamily
T, member 1; K04257 olfactory receptor
Length=369
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 17/32 (53%), Gaps = 1/32 (3%)
Query 1 RQFCWFFIYGIDLFGGILGNYGVTETGMSLPF 32
R+ CW I G FGG L + +T MS PF
Sbjct 188 RRVCWMIIAG-SWFGGSLDGFLLTPITMSFPF 218
> mmu:211978 Zfyve26, 4930465A13, 9330197E15Rik, A630028O16Rik,
Gm893, KIAA0321, mKIAA0321; zinc finger, FYVE domain containing
26
Length=2529
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 14/22 (63%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 83 CSSLSAALLPSLSSPPQCVTVK 104
CSSL+A LL SLSS P V V+
Sbjct 1150 CSSLAAVLLRSLSSDPDHVEVR 1171
Lambda K H
0.323 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072286120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40