bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5219_orf2
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
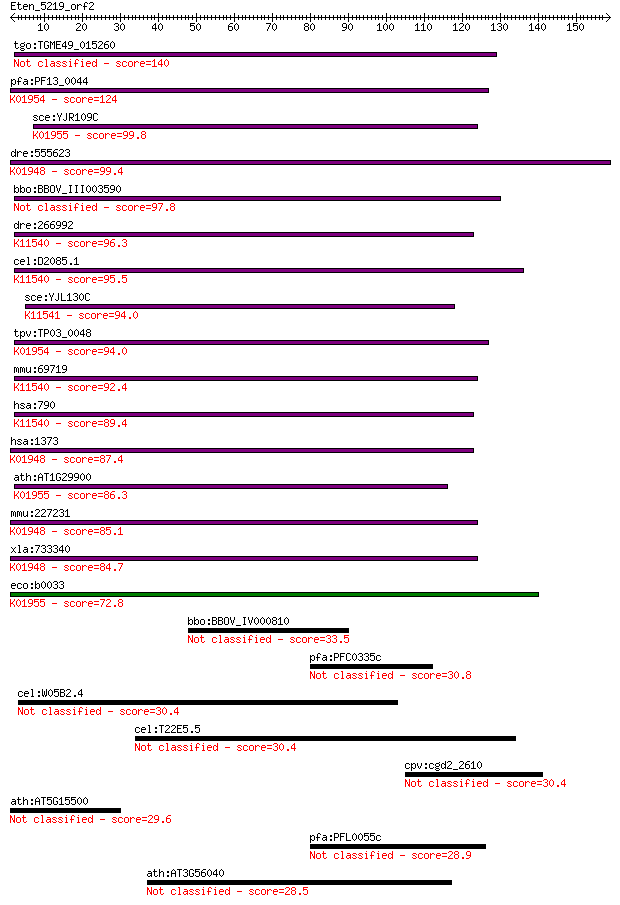
tgo:TGME49_015260 carbamoyl phosphate synthetase II (EC:6.3.4.... 140 2e-33
pfa:PF13_0044 cpsSII; carbamoyl phosphate synthetase (EC:6.3.5... 124 1e-28
sce:YJR109C CPA2; Cpa2p (EC:6.3.5.5); K01955 carbamoyl-phospha... 99.8 3e-21
dre:555623 cps1, si:dkey-225d17.3; carbamoyl-phosphate synthas... 99.4 4e-21
bbo:BBOV_III003590 17.m07338; carbamoyl phosphate synthetase (... 97.8 1e-20
dre:266992 cad, cb456, si:dkey-221h15.3, wu:fc30c12, wu:fc33d0... 96.3 3e-20
cel:D2085.1 pyr-1; PYRimidine biosynthesis family member (pyr-... 95.5 7e-20
sce:YJL130C URA2; Bifunctional carbamoylphosphate synthetase (... 94.0 2e-19
tpv:TP03_0048 glutamine-dependent carbamoyl phosphate synthase... 94.0 2e-19
mmu:69719 Cad, 2410008J01Rik, AU018859, Cpad; carbamoyl-phosph... 92.4 5e-19
hsa:790 CAD; carbamoyl-phosphate synthetase 2, aspartate trans... 89.4 4e-18
hsa:1373 CPS1, CPSASE1; carbamoyl-phosphate synthase 1, mitoch... 87.4 2e-17
ath:AT1G29900 CARB; CARB (CARBAMOYL PHOSPHATE SYNTHETASE B); A... 86.3 4e-17
mmu:227231 Cps1, 4732433M03Rik, CPS, D1Ucla3; carbamoyl-phosph... 85.1 9e-17
xla:733340 cps1; carbamoyl-phosphate synthase 1, mitochondrial... 84.7 1e-16
eco:b0033 carB, cap, ECK0034, JW0031, pyrA; carbamoyl-phosphat... 72.8 5e-13
bbo:BBOV_IV000810 21.m02911; ATP-dependent helicase 33.5 0.32
pfa:PFC0335c conserved Plasmodium protein, unknown function 30.8 1.7
cel:W05B2.4 hypothetical protein 30.4 2.4
cel:T22E5.5 mup-2; MUscle Positioning family member (mup-2) 30.4 2.8
cpv:cgd2_2610 hypothetical protein 30.4 2.8
ath:AT5G15500 ankyrin repeat family protein 29.6 4.7
pfa:PFL0055c RESA-like protein with PHIST and DnaJ domains 28.9 7.8
ath:AT3G56040 UGP3; UGP3 (UDP-GLUCOSE PYROPHOSPHORYLASE 3) 28.5 9.3
> tgo:TGME49_015260 carbamoyl phosphate synthetase II (EC:6.3.4.16
6.3.4.13)
Length=1699
Score = 140 bits (353), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 70/127 (55%), Positives = 88/127 (69%), Gaps = 1/127 (0%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDRQ 61
A G++V+ LHE T ID WFLSKL++I +K L++++ LT AD FY+KKYGFSDRQ
Sbjct 948 AFQLGWTVDALHEKTKIDKWFLSKLQNINDIKRQLTQLTLDDLTRADFFYIKKYGFSDRQ 1007
Query 62 IASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVDD 121
IA Y+ P +++ DV R R L V P +K+IDTLAAEFPA TNYLYLTY G+ DD
Sbjct 1008 IAQYLMNSP-SAAALSQFDVRRRRLHLGVRPSVKQIDTLAAEFPAHTNYLYLTYQGIDDD 1066
Query 122 VEPLNTT 128
V PL T
Sbjct 1067 VSPLAAT 1073
> pfa:PF13_0044 cpsSII; carbamoyl phosphate synthetase (EC:6.3.5.5);
K01954 carbamoyl-phosphate synthase [EC:6.3.5.5]
Length=2375
Score = 124 bits (311), Expect = 1e-28, Method: Composition-based stats.
Identities = 65/135 (48%), Positives = 83/135 (61%), Gaps = 9/135 (6%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDR 60
+A ++++HELTHID WFL K +I L+ L + QL+ DL Y KK+GFSD+
Sbjct 1105 QAFHLNMPMDKIHELTHIDYWFLHKFYNIYNLQNKLKTLKLEQLSFNDLKYFKKHGFSDK 1164
Query 61 QIASYVGGGP---------VEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYL 111
QIA Y+ + RVTE DV +YR+ L + P+IK IDTL+AEFPA TNYL
Sbjct 1165 QIAHYLSFNTSDNNNNNNNISSCRVTENDVMKYREKLGLFPHIKVIDTLSAEFPALTNYL 1224
Query 112 YLTYHGVVDDVEPLN 126
YLTY G DV PLN
Sbjct 1225 YLTYQGQEHDVLPLN 1239
> sce:YJR109C CPA2; Cpa2p (EC:6.3.5.5); K01955 carbamoyl-phosphate
synthase large subunit [EC:6.3.5.5]
Length=1118
Score = 99.8 bits (247), Expect = 3e-21, Method: Composition-based stats.
Identities = 55/118 (46%), Positives = 72/118 (61%), Gaps = 3/118 (2%)
Query 7 YSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFY-VKKYGFSDRQIASY 65
Y+VER++EL+ ID WFL K +I + + L V S DL KK GFSD+QIA
Sbjct 460 YTVERVNELSKIDKWFLYKCMNIVNIYKELESVKSLSDLSKDLLQRAKKLGFSDKQIAVT 519
Query 66 VGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVDDVE 123
+ + E ++ RK L + P++KRIDTLAAEFPAQTNYLY TY+ +DVE
Sbjct 520 INKHA--STNINELEIRSLRKTLGIIPFVKRIDTLAAEFPAQTNYLYTTYNATKNDVE 575
> dre:555623 cps1, si:dkey-225d17.3; carbamoyl-phosphate synthase
1, mitochondrial; K01948 carbamoyl-phosphate synthase (ammonia)
[EC:6.3.4.16]
Length=1524
Score = 99.4 bits (246), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 56/158 (35%), Positives = 87/158 (55%), Gaps = 8/158 (5%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDR 60
+AL G +V+++H+LT ID WFL KL+HI ++++ L + A + L K GFSDR
Sbjct 846 QALHSGVTVDQIHDLTAIDKWFLHKLKHITEMEQRLGQYKSATVPRDLLLKAKMDGFSDR 905
Query 61 QIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVD 120
Q+ + + ++E + R + P++K+IDTLAAE+PA TNYLY TYHG
Sbjct 906 QVG--------QAMDISEGEARVLRLNQNIRPWVKQIDTLAAEYPAATNYLYCTYHGQEH 957
Query 121 DVEPLNTTDLPPVLGPWGGGHQESAQFTAPSAQRQQSQ 158
D++ + + GP+ G + A S+ R Q
Sbjct 958 DLDFKDHGTMVVGCGPYHIGSSVEFDWCAVSSIRALRQ 995
> bbo:BBOV_III003590 17.m07338; carbamoyl phosphate synthetase
(EC:6.3.5.5)
Length=1632
Score = 97.8 bits (242), Expect = 1e-20, Method: Composition-based stats.
Identities = 58/129 (44%), Positives = 78/129 (60%), Gaps = 6/129 (4%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKV-SPAQLTSADLFYVKKYGFSDR 60
A G +V +H LT IDPWFL +L H+ L LS + S + T A + Y K YGFSDR
Sbjct 938 AFELGMTVSDIHGLTKIDPWFLHRLHHLHILNAHLSTLPSLSSFTPAMMRYYKVYGFSDR 997
Query 61 QIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVD 120
QI+ + V +E+DV RK+ + P++K IDT+AAE+PA+TNY YLTY+G+
Sbjct 998 QISREIVKSTV-----SEDDVRELRKSWGIVPFVKVIDTMAAEYPAKTNYCYLTYNGIES 1052
Query 121 DVEPLNTTD 129
DV P D
Sbjct 1053 DVLPCGPID 1061
> dre:266992 cad, cb456, si:dkey-221h15.3, wu:fc30c12, wu:fc33d01,
wu:fc67g02; carbamoyl-phosphate synthetase 2, aspartate
transcarbamylase, and dihydroorotase (EC:2.1.3.2); K11540 carbamoyl-phosphate
synthase / aspartate carbamoyltransferase
/ dihydroorotase [EC:6.3.5.5 2.1.3.2 3.5.2.3]
Length=2230
Score = 96.3 bits (238), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 56/123 (45%), Positives = 77/123 (62%), Gaps = 10/123 (8%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLS--KVSPAQLTSADLFYVKKYGFSD 59
AL GY+VERL+ELT ID WFL K+++I K+ L K + + + K+ GFSD
Sbjct 819 ALHAGYTVERLYELTKIDHWFLHKMKNIADHKKLLETYKQDESAMPPEIMRKAKQLGFSD 878
Query 60 RQIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVV 119
+QIA + ++ TE V + R+ + P +K+IDT+AAE+PAQTNYLYLTYHG
Sbjct 879 KQIA--------QAVQSTELAVRKLRRDWKIFPVVKQIDTVAAEWPAQTNYLYLTYHGSE 930
Query 120 DDV 122
DV
Sbjct 931 SDV 933
> cel:D2085.1 pyr-1; PYRimidine biosynthesis family member (pyr-1);
K11540 carbamoyl-phosphate synthase / aspartate carbamoyltransferase
/ dihydroorotase [EC:6.3.5.5 2.1.3.2 3.5.2.3]
Length=2198
Score = 95.5 bits (236), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 56/139 (40%), Positives = 78/139 (56%), Gaps = 14/139 (10%)
Query 2 ALARG-----YSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYG 56
ALARG + VE+ HELT ID WFL ++++I + L K +++ L K+ G
Sbjct 812 ALARGMYYGDFDVEKAHELTRIDRWFLFRMQNIVDIYHRLEKTDVNTVSAELLLEAKQAG 871
Query 57 FSDRQIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYH 116
FSDRQIA +G E V R + P +K+IDT+A E+PAQTNYLY T++
Sbjct 872 FSDRQIAKKIGS--------NEYTVREARFVKGITPCVKQIDTVAGEWPAQTNYLYTTFN 923
Query 117 GVVDDVEPLNTTDLPPVLG 135
G+ +DV N + VLG
Sbjct 924 GIENDVS-FNMKNAVMVLG 941
> sce:YJL130C URA2; Bifunctional carbamoylphosphate synthetase
(CPSase)-aspartate transcarbamylase (ATCase), catalyzes the
first two enzymatic steps in the de novo biosynthesis of pyrimidines;
both activities are subject to feedback inhibition
by UTP (EC:6.3.5.5 2.1.3.2); K11541 carbamoyl-phosphate synthase
/ aspartate carbamoyltransferase [EC:6.3.5.5 2.1.3.2]
Length=2214
Score = 94.0 bits (232), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 49/114 (42%), Positives = 70/114 (61%), Gaps = 9/114 (7%)
Query 5 RGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKV-SPAQLTSADLFYVKKYGFSDRQIA 63
+GYSV+++ E+T ID WFL+KL + Q E +S + +L S L K+ GF DRQIA
Sbjct 865 KGYSVDKVWEMTRIDKWFLNKLHDLVQFAEKISSFGTKEELPSLVLRQAKQLGFDDRQIA 924
Query 64 SYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHG 117
++ V + R RK + P++K+IDT+AAEFPA TNYLY+TY+
Sbjct 925 RFLDSNEVA--------IRRLRKEYGITPFVKQIDTVAAEFPAYTNYLYMTYNA 970
> tpv:TP03_0048 glutamine-dependent carbamoyl phosphate synthase
(EC:6.3.5.5); K01954 carbamoyl-phosphate synthase [EC:6.3.5.5]
Length=1680
Score = 94.0 bits (232), Expect = 2e-19, Method: Composition-based stats.
Identities = 54/133 (40%), Positives = 79/133 (59%), Gaps = 8/133 (6%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVS-PAQLTSADLFYVKKYGFSDR 60
A G +V++++E T ID WFL++L +I E L K++ +LT + L Y K GFSDR
Sbjct 942 AFELGITVQQINEFTRIDNWFLNRLHNIYLCSERLKKLTRKEELTHSQLLYYKVLGFSDR 1001
Query 61 QIASYVG--GGPVEGLRVTE----EDVWR-YRKALVVEPYIKRIDTLAAEFPAQTNYLYL 113
Q++ + G V + +E E+ +R YR L+V P + IDTLAAE+P TNY Y+
Sbjct 1002 QMSLLINNKGNVVNLVNKSELEKYENEFRDYRLKLLVTPKVNIIDTLAAEYPVVTNYCYM 1061
Query 114 TYHGVVDDVEPLN 126
TY+ D+ PLN
Sbjct 1062 TYNSTEHDIRPLN 1074
> mmu:69719 Cad, 2410008J01Rik, AU018859, Cpad; carbamoyl-phosphate
synthetase 2, aspartate transcarbamylase, and dihydroorotase
(EC:2.1.3.2); K11540 carbamoyl-phosphate synthase / aspartate
carbamoyltransferase / dihydroorotase [EC:6.3.5.5 2.1.3.2
3.5.2.3]
Length=2225
Score = 92.4 bits (228), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 58/126 (46%), Positives = 75/126 (59%), Gaps = 16/126 (12%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLE----HIQQLKESLSKVSPAQLTSADLFYVKKYGF 57
AL GYSVERL+ELT ID WFL +++ H Q L++ + P L L K GF
Sbjct 816 ALWAGYSVERLYELTRIDCWFLHRMKRIVTHAQLLEQHRGQALPQDL----LHQAKCLGF 871
Query 58 SDRQIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHG 117
SD+QIA V TE V + R+ L + P +K+IDT+AAE+PAQTNYLYLTY G
Sbjct 872 SDKQIALAV--------LSTELAVRKLRQELGICPAVKQIDTVAAEWPAQTNYLYLTYWG 923
Query 118 VVDDVE 123
D++
Sbjct 924 NTHDLD 929
> hsa:790 CAD; carbamoyl-phosphate synthetase 2, aspartate transcarbamylase,
and dihydroorotase (EC:2.1.3.2 3.5.2.3 6.3.5.5);
K11540 carbamoyl-phosphate synthase / aspartate carbamoyltransferase
/ dihydroorotase [EC:6.3.5.5 2.1.3.2 3.5.2.3]
Length=2225
Score = 89.4 bits (220), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 57/125 (45%), Positives = 74/125 (59%), Gaps = 16/125 (12%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLE----HIQQLKESLSKVSPAQLTSADLFYVKKYGF 57
AL GYSV+RL+ELT ID WFL +++ H Q L++ + P L L K GF
Sbjct 816 ALWAGYSVDRLYELTRIDRWFLHRMKRIIAHAQLLEQHRGQPLPPDL----LQQAKCLGF 871
Query 58 SDRQIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHG 117
SD+QIA V TE V + R+ L + P +K+IDT+AAE+PAQTNYLYLTY G
Sbjct 872 SDKQIALAV--------LSTELAVRKLRQELGICPAVKQIDTVAAEWPAQTNYLYLTYWG 923
Query 118 VVDDV 122
D+
Sbjct 924 TTHDL 928
> hsa:1373 CPS1, CPSASE1; carbamoyl-phosphate synthase 1, mitochondrial
(EC:6.3.4.16); K01948 carbamoyl-phosphate synthase
(ammonia) [EC:6.3.4.16]
Length=1506
Score = 87.4 bits (215), Expect = 2e-17, Method: Composition-based stats.
Identities = 45/122 (36%), Positives = 73/122 (59%), Gaps = 8/122 (6%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDR 60
+A+ S++ + +LT+ID WFL K+ I ++++L ++ +T L K+ GFSD+
Sbjct 862 KAIDDNMSLDEIEKLTYIDKWFLYKMRDILNMEKTLKGLNSESMTEETLKRAKEIGFSDK 921
Query 61 QIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVD 120
QI+ +G +TE R + P++K+IDTLAAE+P+ TNYLY+TY+G
Sbjct 922 QISKCLG--------LTEAQTRELRLKKNIHPWVKQIDTLAAEYPSVTNYLYVTYNGQEH 973
Query 121 DV 122
DV
Sbjct 974 DV 975
> ath:AT1G29900 CARB; CARB (CARBAMOYL PHOSPHATE SYNTHETASE B);
ATP binding / carbamoyl-phosphate synthase/ catalytic (EC:6.3.4.16);
K01955 carbamoyl-phosphate synthase large subunit
[EC:6.3.5.5]
Length=1187
Score = 86.3 bits (212), Expect = 4e-17, Method: Composition-based stats.
Identities = 44/114 (38%), Positives = 71/114 (62%), Gaps = 8/114 (7%)
Query 2 ALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDRQ 61
A+ +G ++ ++EL+ +D WFL++L+ + +++ L + +++T DL+ VKK GFSD+Q
Sbjct 533 AMKKGMKIDEIYELSMVDKWFLTQLKELVDVEQYLMSGTLSEITKEDLYEVKKRGFSDKQ 592
Query 62 IASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTY 115
IA + TEE+V R +L V P KR+DT AAEF A T Y+Y +Y
Sbjct 593 IAF--------ATKTTEEEVRTKRISLGVVPSYKRVDTCAAEFEAHTPYMYSSY 638
> mmu:227231 Cps1, 4732433M03Rik, CPS, D1Ucla3; carbamoyl-phosphate
synthetase 1 (EC:6.3.4.16); K01948 carbamoyl-phosphate
synthase (ammonia) [EC:6.3.4.16]
Length=1500
Score = 85.1 bits (209), Expect = 9e-17, Method: Composition-based stats.
Identities = 45/123 (36%), Positives = 71/123 (57%), Gaps = 8/123 (6%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDR 60
+AL S++ + LT ID WFL K+ I + ++L ++ +T L K+ GFSD+
Sbjct 856 KALENNMSLDEIVRLTSIDKWFLYKMRDILNMDKTLKGLNSDSVTEETLRKAKEIGFSDK 915
Query 61 QIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVD 120
QI+ +G +TE R + P++K+IDTLAAE+P+ TNYLY+TY+G
Sbjct 916 QISKCLG--------LTEAQTRELRLKKNIHPWVKQIDTLAAEYPSVTNYLYVTYNGQEH 967
Query 121 DVE 123
D++
Sbjct 968 DIK 970
> xla:733340 cps1; carbamoyl-phosphate synthase 1, mitochondrial;
K01948 carbamoyl-phosphate synthase (ammonia) [EC:6.3.4.16]
Length=1494
Score = 84.7 bits (208), Expect = 1e-16, Method: Composition-based stats.
Identities = 45/123 (36%), Positives = 67/123 (54%), Gaps = 8/123 (6%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDR 60
+ L G ++ +H+LT ID WFL K++ I ++ +L + L K+ GFSDR
Sbjct 850 KGLETGIHIDEIHKLTSIDKWFLYKMQDILNMENTLRSTRSESIPEETLRRAKQIGFSDR 909
Query 61 QIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVD 120
I +G ++E R P++K+IDTLAAE+PA TNYLYLTY+G
Sbjct 910 YIGKCLG--------LSEAQTRELRLNKNFTPWVKQIDTLAAEYPAITNYLYLTYNGQEH 961
Query 121 DVE 123
D++
Sbjct 962 DIK 964
> eco:b0033 carB, cap, ECK0034, JW0031, pyrA; carbamoyl-phosphate
synthase large subunit (EC:6.3.5.5); K01955 carbamoyl-phosphate
synthase large subunit [EC:6.3.5.5]
Length=1073
Score = 72.8 bits (177), Expect = 5e-13, Method: Composition-based stats.
Identities = 45/139 (32%), Positives = 74/139 (53%), Gaps = 11/139 (7%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHIQQLKESLSKVSPAQLTSADLFYVKKYGFSDR 60
+A G SV+ + LT+ID WFL ++E + +L+E +++V L + L +K+ GF+D
Sbjct 441 DAFRAGLSVDGVFNLTNIDRWFLVQIEELVRLEEKVAEVGITGLNADFLRQLKRKGFADA 500
Query 61 QIASYVGGGPVEGLRVTEEDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVD 120
++A G V E ++ + R + P KR+DT AAEF T Y+Y TY +
Sbjct 501 RLAKLAG--------VREAEIRKLRDQYDLHPVYKRVDTCAAEFATDTAYMYSTYE---E 549
Query 121 DVEPLNTTDLPPVLGPWGG 139
+ E +TD ++ GG
Sbjct 550 ECEANPSTDREKIMVLGGG 568
> bbo:BBOV_IV000810 21.m02911; ATP-dependent helicase
Length=706
Score = 33.5 bits (75), Expect = 0.32, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 48 DLFYVKKYGFSDRQIASYVGGGPVEGLRVTEEDVWRYRKALV 89
D+ YV GFS R++A Y+ G VE ++ D R R L+
Sbjct 317 DVAYVVDCGFSRRRVADYINTGTVETKVISTRDEMRQRSNLI 358
> pfa:PFC0335c conserved Plasmodium protein, unknown function
Length=3724
Score = 30.8 bits (68), Expect = 1.7, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 80 DVWRYRKALVVEPYIKRIDTLAAEFPAQTNYL 111
D+ RYR+ L + +KRID + A+F NY+
Sbjct 3082 DILRYRQNLYHQLNMKRIDKICAQFLRDNNYI 3113
> cel:W05B2.4 hypothetical protein
Length=2265
Score = 30.4 bits (67), Expect = 2.4, Method: Composition-based stats.
Identities = 31/117 (26%), Positives = 48/117 (41%), Gaps = 17/117 (14%)
Query 3 LARGYSVERL---HELTHIDPWFLSKLEHIQQLKESLSKVSPAQL-------TSADLFYV 52
L +VE L HE + P F + + ++ S PAQL +S +
Sbjct 1242 LPTNIAVEHLGVFHEFVEVSPAFKFETSPLSSVRTSSIPKLPAQLVSTGQPPSSPHSVLL 1301
Query 53 KKYG--FSDRQIASYVGGGPVEGLRVT-----EEDVWRYRKALVVEPYIKRIDTLAA 102
+K G +R+I+ VG E L+ T E+ +W Y V + + R D AA
Sbjct 1302 EKIGAKIRNREISGCVGSLAKETLKETLQNFPEKIIWVYTDVFVRDQMVTRSDEFAA 1358
> cel:T22E5.5 mup-2; MUscle Positioning family member (mup-2)
Length=405
Score = 30.4 bits (67), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 49/115 (42%), Gaps = 19/115 (16%)
Query 34 ESLSKVSPAQLTSADLF--YVKKYGFSDRQI----------ASYVGGG---PVEGLRVTE 78
E+ S V P ++T+A F + + DR+ AS V G P E R
Sbjct 288 EAASSVHPPKITTASKFDRQTDRRSYGDRRYLFENPEEEKQASLVRGTGRPPAEWGRKQN 347
Query 79 EDVWRYRKALVVEPYIKRIDTLAAEFPAQTNYLYLTYHGVVDDVEPLNTTDLPPV 133
E++ + RK L Y++++ A P + L++ DD EP T+ P V
Sbjct 348 EELEQIRKNLEPPKYVEQVKVEGARAPVEPVPLFVP----TDDFEPQQVTEDPNV 398
> cpv:cgd2_2610 hypothetical protein
Length=2612
Score = 30.4 bits (67), Expect = 2.8, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 20/37 (54%), Gaps = 1/37 (2%)
Query 105 PAQTNYLYLTYHGVVDDV-EPLNTTDLPPVLGPWGGG 140
P T Y + Y+ +V D+ +P+ T + P P+ GG
Sbjct 674 PMNTYYYFQHYYTIVTDIYQPIQLTQITPYTDPYAGG 710
> ath:AT5G15500 ankyrin repeat family protein
Length=457
Score = 29.6 bits (65), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 12/29 (41%), Positives = 20/29 (68%), Gaps = 0/29 (0%)
Query 1 EALARGYSVERLHELTHIDPWFLSKLEHI 29
EA A+ +++ L+EL H DP+ L K +H+
Sbjct 7 EAAAKSGNIDLLYELIHEDPYVLDKTDHV 35
> pfa:PFL0055c RESA-like protein with PHIST and DnaJ domains
Length=900
Score = 28.9 bits (63), Expect = 7.8, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 80 DVWRYRKALVVEPYIKRIDTLAAEFPAQTNYL-YLTYHGVVDDVEPL 125
+VW Y+KAL + Y K +++ + A +N+L Y ++ D+E +
Sbjct 264 NVWYYKKALYMNEYRKLVNSCRIAWKALSNHLKYTLKMTIIYDIEKM 310
> ath:AT3G56040 UGP3; UGP3 (UDP-GLUCOSE PYROPHOSPHORYLASE 3)
Length=883
Score = 28.5 bits (62), Expect = 9.3, Method: Composition-based stats.
Identities = 26/94 (27%), Positives = 36/94 (38%), Gaps = 18/94 (19%)
Query 37 SKVSPAQLTSADLFYVKKYGFSDRQIASYVGGGPVEGLRVTEEDV-----WRYRKALVVE 91
+ V+ L L Y KK GF+ + G EG+ V E W Y + +
Sbjct 468 TDVTLLALAGIGLRYNKKLGFA----SCKRNAGATEGINVLMEKKNFDGKWEYGISCIEY 523
Query 92 PYIKRIDT---------LAAEFPAQTNYLYLTYH 116
+ D L A+FPA TN LY+ H
Sbjct 524 TEFDKFDISNRSPSSNGLQADFPANTNILYVDLH 557
Lambda K H
0.316 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3582056500
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40