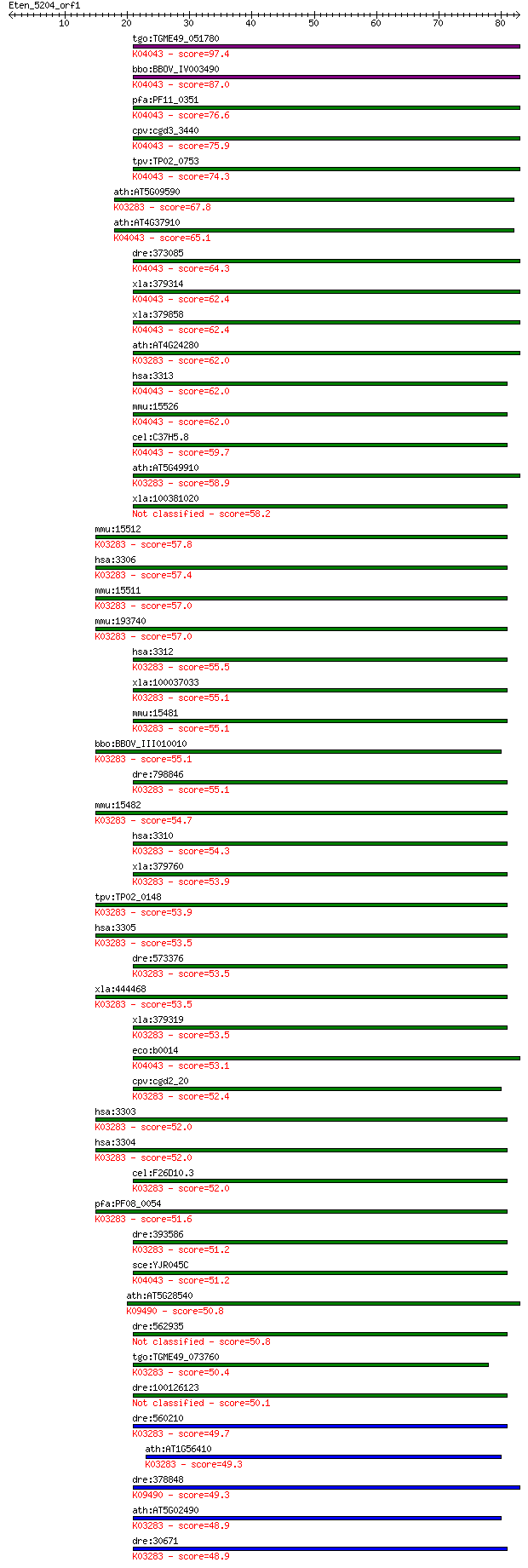
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5204_orf1
Length=82
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_051780 heat shock protein 70, putative ; K04043 mol... 97.4 9e-21
bbo:BBOV_IV003490 21.m02799; heat shock protein 70; K04043 mol... 87.0 1e-17
pfa:PF11_0351 heat shock protein hsp70 homologue; K04043 molec... 76.6 2e-14
cpv:cgd3_3440 heat shock protein HSP70, mitochondrial ; K04043... 75.9 3e-14
tpv:TP02_0753 heat shock protein 70; K04043 molecular chaperon... 74.3 8e-14
ath:AT5G09590 MTHSC70-2 (MITOCHONDRIAL HSP70 2); ATP binding; ... 67.8 8e-12
ath:AT4G37910 mtHsc70-1 (mitochondrial heat shock protein 70-1... 65.1 5e-11
dre:373085 hspa9, MGC86654, cb740, crs, hspa9b, wu:fc14d08, wu... 64.3 9e-11
xla:379314 hspa9-a, MGC52616, csa, grp-75, grp75, hspa9, hspa9... 62.4 3e-10
xla:379858 hspa9-b, MGC53528, csa, grp-75, grp75, hspa9b, mot,... 62.4 4e-10
ath:AT4G24280 cpHsc70-1 (chloroplast heat shock protein 70-1);... 62.0 4e-10
hsa:3313 HSPA9, CSA, GRP-75, GRP75, HSPA9B, MGC4500, MOT, MOT2... 62.0 4e-10
mmu:15526 Hspa9, 74kDa, Csa, Grp75, Hsc74, Hsp74, Hsp74a, Hspa... 62.0 5e-10
cel:C37H5.8 hsp-6; Heat Shock Protein family member (hsp-6); K... 59.7 2e-09
ath:AT5G49910 CPHSC70-2EAT SHOCK PROTEIN 70-2 (CHLOROPLAST HEA... 58.9 4e-09
xla:100381020 hspa1a, hsp70-1, hsp70-1a, hsp70i, hsp72, hspa1,... 58.2 6e-09
mmu:15512 Hspa2, 70kDa, HSP70.2, HSP70A2, Hsp70-2, MGC58299, M... 57.8 8e-09
hsa:3306 HSPA2, HSP70-2, HSP70-3; heat shock 70kDa protein 2; ... 57.4 1e-08
mmu:15511 Hspa1b, Hsp70, Hsp70-1, Hsp70.1, hsp68; heat shock p... 57.0 1e-08
mmu:193740 Hspa1a, Hsp70-3, Hsp70.3, Hsp72, MGC189852, hsp68, ... 57.0 1e-08
hsa:3312 HSPA8, HSC54, HSC70, HSC71, HSP71, HSP73, HSPA10, LAP... 55.5 5e-08
xla:100037033 hspa1b; heat shock 70kDa protein 1B; K03283 heat... 55.1 5e-08
mmu:15481 Hspa8, 2410008N15Rik, Hsc70, Hsc71, Hsc73, Hsp73, Hs... 55.1 5e-08
bbo:BBOV_III010010 17.m07869; dnaK protein; K03283 heat shock ... 55.1 5e-08
dre:798846 novel protein similar to vertebrate heat shock 70kD... 55.1 6e-08
mmu:15482 Hspa1l, Hsc70t, MGC150263, MGC150264, Msh5; heat sho... 54.7 6e-08
hsa:3310 HSPA6; heat shock 70kDa protein 6 (HSP70B'); K03283 h... 54.3 9e-08
xla:379760 hspa1l, MGC52655, hsc70, hsc70.I; heat shock 70kDa ... 53.9 1e-07
tpv:TP02_0148 heat shock protein 70; K03283 heat shock 70kDa p... 53.9 1e-07
hsa:3305 HSPA1L, HSP70-1L, HSP70-HOM, HSP70T, hum70t; heat sho... 53.5 2e-07
dre:573376 hspa8, MGC55272, hsc70, wu:fb01g06, wu:fi48b06; hea... 53.5 2e-07
xla:444468 hspa2, MGC81782; heat shock 70kDa protein 2; K03283... 53.5 2e-07
xla:379319 hspa8, MGC53952, hsc54, hsc70, hsc71, hsp71, hsp73,... 53.5 2e-07
eco:b0014 dnaK, ECK0014, groPAB, groPC, groPF, grpC, grpF, JW0... 53.1 2e-07
cpv:cgd2_20 heat shock 70 (HSP70) protein ; K03283 heat shock ... 52.4 4e-07
hsa:3303 HSPA1A, FLJ54303, FLJ54370, FLJ54392, FLJ54408, FLJ75... 52.0 4e-07
hsa:3304 HSPA1B, FLJ54328, HSP70-1B, HSP70-2, HSPA1A; heat sho... 52.0 4e-07
cel:F26D10.3 hsp-1; Heat Shock Protein family member (hsp-1); ... 52.0 5e-07
pfa:PF08_0054 heat shock 70 kDa protein; K03283 heat shock 70k... 51.6 6e-07
dre:393586 MGC63663; zgc:63663; K03283 heat shock 70kDa protei... 51.2 7e-07
sce:YJR045C SSC1, ENS1; Hsp70 family ATPase, constituent of th... 51.2 9e-07
ath:AT5G28540 BIP1; BIP1; ATP binding; K09490 heat shock 70kDa... 50.8 9e-07
dre:562935 heat shock cognate 70 kDa protein 50.8 1e-06
tgo:TGME49_073760 heat shock protein 70, putative ; K03283 hea... 50.4 1e-06
dre:100126123 zgc:174006 50.1 2e-06
dre:560210 hsp70l, hsp70-4; heat shock cognate 70-kd protein, ... 49.7 2e-06
ath:AT1G56410 ERD2; ERD2 (EARLY-RESPONSIVE TO DEHYDRATION 2); ... 49.3 3e-06
dre:378848 hspa5, cb865, fb60h09, fi36d04, wu:fb60h09, wu:fi36... 49.3 3e-06
ath:AT5G02490 heat shock cognate 70 kDa protein 2 (HSC70-2) (H... 48.9 3e-06
dre:30671 hsp70, hsp70-4, hspa1a; heat shock cognate 70-kd pro... 48.9 4e-06
> tgo:TGME49_051780 heat shock protein 70, putative ; K04043 molecular
chaperone DnaK
Length=728
Score = 97.4 bits (241), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 47/62 (75%), Positives = 59/62 (95%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
AVDKSTGKRQ+ITIQSSGGLS++QI+QMV+DAE +K++D+++KD V AKNEAETL+YSVE
Sbjct 588 AVDKSTGKRQEITIQSSGGLSDSQIEQMVKDAEMYKEQDEKKKDAVQAKNEAETLIYSVE 647
Query 81 KQ 82
KQ
Sbjct 648 KQ 649
> bbo:BBOV_IV003490 21.m02799; heat shock protein 70; K04043 molecular
chaperone DnaK
Length=654
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 43/62 (69%), Positives = 55/62 (88%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
AVDKSTG++Q+ITIQSSGGLS+ Q+++MV+DAE FK D+++K LV A+NEAETL YSVE
Sbjct 517 AVDKSTGRKQEITIQSSGGLSDEQVERMVKDAEAFKQSDEQRKLLVDARNEAETLCYSVE 576
Query 81 KQ 82
KQ
Sbjct 577 KQ 578
> pfa:PF11_0351 heat shock protein hsp70 homologue; K04043 molecular
chaperone DnaK
Length=663
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/62 (61%), Positives = 53/62 (85%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A+DK T K+QQITIQSSGGLS+ +I++MV++AE +++DQ +K+L +KNEAETL+YS E
Sbjct 522 AIDKMTNKKQQITIQSSGGLSKEEIEKMVQEAELNREKDQLKKNLTDSKNEAETLIYSSE 581
Query 81 KQ 82
KQ
Sbjct 582 KQ 583
> cpv:cgd3_3440 heat shock protein HSP70, mitochondrial ; K04043
molecular chaperone DnaK
Length=683
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 34/62 (54%), Positives = 50/62 (80%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A+DKSTGK+ +ITIQSSGGLS A+I++M+ +AE ++ DQ +K+L+ KN+AE +YSV+
Sbjct 532 AIDKSTGKKHEITIQSSGGLSGAEIEKMIREAEEYRANDQAKKELIDLKNDAEAFIYSVQ 591
Query 81 KQ 82
Q
Sbjct 592 NQ 593
> tpv:TP02_0753 heat shock protein 70; K04043 molecular chaperone
DnaK
Length=681
Score = 74.3 bits (181), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 40/62 (64%), Positives = 56/62 (90%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
AVDKSTGKRQ+ITIQSSGGLSE ++++MV++A +K++D R+K+LV +NE+E+L+YSVE
Sbjct 538 AVDKSTGKRQEITIQSSGGLSEEEVEKMVKEASNYKEQDDRRKELVDVRNESESLLYSVE 597
Query 81 KQ 82
KQ
Sbjct 598 KQ 599
> ath:AT5G09590 MTHSC70-2 (MITOCHONDRIAL HSP70 2); ATP binding;
K03283 heat shock 70kDa protein 1/8
Length=682
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 35/64 (54%), Positives = 48/64 (75%), Gaps = 0/64 (0%)
Query 18 TDRAVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVY 77
T A DK+TGK QQITI+SSGGLSE I++MV +AE +D+ +K+L+ KN A+T +Y
Sbjct 534 TVSAKDKTTGKVQQITIRSSGGLSEDDIQKMVREAELHAQKDKERKELIDTKNTADTTIY 593
Query 78 SVEK 81
S+EK
Sbjct 594 SIEK 597
> ath:AT4G37910 mtHsc70-1 (mitochondrial heat shock protein 70-1);
ATP binding; K04043 molecular chaperone DnaK
Length=682
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 34/64 (53%), Positives = 47/64 (73%), Gaps = 0/64 (0%)
Query 18 TDRAVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVY 77
T A DK+TGK Q ITI+SSGGLS+ +I +MV++AE +DQ +K L+ +N A+T +Y
Sbjct 529 TVSAKDKATGKEQNITIRSSGGLSDDEINRMVKEAELNAQKDQEKKQLIDLRNSADTTIY 588
Query 78 SVEK 81
SVEK
Sbjct 589 SVEK 592
> dre:373085 hspa9, MGC86654, cb740, crs, hspa9b, wu:fc14d08,
wu:fc27c10, wu:fc38a06; heat shock protein 9; K04043 molecular
chaperone DnaK
Length=682
Score = 64.3 bits (155), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 33/62 (53%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A DK TG+ QQI IQSSGGLS+ I+ MV++AE++ +ED+R+KD V A N AE +V+ E
Sbjct 537 AKDKGTGREQQIVIQSSGGLSKDDIENMVKNAEKYAEEDRRRKDRVEAVNMAEGIVHDTE 596
Query 81 KQ 82
+
Sbjct 597 SK 598
> xla:379314 hspa9-a, MGC52616, csa, grp-75, grp75, hspa9, hspa9b,
mot, mot2, pbp74; heat shock 70kDa protein 9 (mortalin);
K04043 molecular chaperone DnaK
Length=670
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A DK TG+ QQI IQSSGGLS+ I+ MV++AE++ +ED+R+K+ V A N AE +++ E
Sbjct 528 AKDKGTGREQQIVIQSSGGLSKDDIENMVKNAEKYAEEDRRRKERVEAVNNAEGIIHDTE 587
Query 81 KQ 82
+
Sbjct 588 SK 589
> xla:379858 hspa9-b, MGC53528, csa, grp-75, grp75, hspa9b, mot,
mot2, pbp74; heat shock 70kDa protein 9 (mortalin); K04043
molecular chaperone DnaK
Length=670
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A DK TG+ QQI IQSSGGLS+ I+ MV++AE++ +ED+R+K+ V A N AE +++ E
Sbjct 528 AKDKGTGREQQIVIQSSGGLSKDDIENMVKNAEKYAEEDRRRKEQVEAVNNAEGIIHDTE 587
Query 81 KQ 82
+
Sbjct 588 SK 589
> ath:AT4G24280 cpHsc70-1 (chloroplast heat shock protein 70-1);
ATP binding; K03283 heat shock 70kDa protein 1/8
Length=718
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 45/62 (72%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
AVDK TGK+Q ITI + L + ++ QMV++AERF +D+ ++D + KN+A+++VY E
Sbjct 559 AVDKGTGKKQDITITGASTLPKDEVDQMVQEAERFAKDDKEKRDAIDTKNQADSVVYQTE 618
Query 81 KQ 82
KQ
Sbjct 619 KQ 620
> hsa:3313 HSPA9, CSA, GRP-75, GRP75, HSPA9B, MGC4500, MOT, MOT2,
MTHSP75, PBP74; heat shock 70kDa protein 9 (mortalin); K04043
molecular chaperone DnaK
Length=679
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 31/60 (51%), Positives = 45/60 (75%), Gaps = 0/60 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A DK TG+ QQI IQSSGGLS+ I+ MV++AE++ +ED+R+K+ V A N AE +++ E
Sbjct 534 AKDKGTGREQQIVIQSSGGLSKDDIENMVKNAEKYAEEDRRKKERVEAVNMAEGIIHDTE 593
> mmu:15526 Hspa9, 74kDa, Csa, Grp75, Hsc74, Hsp74, Hsp74a, Hspa9a,
Mortalin, Mot-2, Mot2, Mthsp70, Pbp74; heat shock protein
9; K04043 molecular chaperone DnaK
Length=679
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 31/60 (51%), Positives = 45/60 (75%), Gaps = 0/60 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A DK TG+ QQI IQSSGGLS+ I+ MV++AE++ +ED+R+K+ V A N AE +++ E
Sbjct 534 AKDKGTGREQQIVIQSSGGLSKDDIENMVKNAEKYAEEDRRKKERVEAVNMAEGIIHDTE 593
> cel:C37H5.8 hsp-6; Heat Shock Protein family member (hsp-6);
K04043 molecular chaperone DnaK
Length=657
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 44/60 (73%), Gaps = 0/60 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A D+ TGK QQI IQSSGGLS+ QI+ M+++AE+ ED ++K+LV N+AE +++ E
Sbjct 513 ARDRGTGKEQQIVIQSSGGLSKDQIENMIKEAEKNAAEDAKRKELVEVINQAEGIIHDTE 572
> ath:AT5G49910 CPHSC70-2EAT SHOCK PROTEIN 70-2 (CHLOROPLAST HEAT
SHOCK PROTEIN 70-2); ATP binding / unfolded protein binding;
K03283 heat shock 70kDa protein 1/8
Length=718
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 43/62 (69%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A DK TGK+Q ITI + L + ++ MV++AERF ED+ ++D + KN+A+++VY E
Sbjct 559 ASDKGTGKKQDITITGASTLPKDEVDTMVQEAERFAKEDKEKRDAIDTKNQADSVVYQTE 618
Query 81 KQ 82
KQ
Sbjct 619 KQ 620
> xla:100381020 hspa1a, hsp70-1, hsp70-1a, hsp70i, hsp72, hspa1,
hspa1b; heat shock 70kDa protein 1A
Length=652
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 48/61 (78%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ +I++MV++A+R+K ED+ Q+D ++AKN E+L +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKDEIERMVQEADRYKTEDEAQRDKISAKNSLESLAFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> mmu:15512 Hspa2, 70kDa, HSP70.2, HSP70A2, Hsp70-2, MGC58299,
MGC7795; heat shock protein 2; K03283 heat shock 70kDa protein
1/8
Length=633
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 46/67 (68%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A DKSTGK +ITI + G LS+ I +MV++AER+K ED+ +D VAAKN E
Sbjct 489 LNVT--AADKSTGKENKITITNDKGRLSKDDIDRMVQEAERYKSEDEANRDRVAAKNAVE 546
Query 74 TLVYSVE 80
+ Y+++
Sbjct 547 SYTYNIK 553
> hsa:3306 HSPA2, HSP70-2, HSP70-3; heat shock 70kDa protein 2;
K03283 heat shock 70kDa protein 1/8
Length=639
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 46/67 (68%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A DKSTGK +ITI + G LS+ I +MV++AER+K ED+ +D VAAKN E
Sbjct 489 LNVT--AADKSTGKENKITITNDKGRLSKDDIDRMVQEAERYKSEDEANRDRVAAKNALE 546
Query 74 TLVYSVE 80
+ Y+++
Sbjct 547 SYTYNIK 553
> mmu:15511 Hspa1b, Hsp70, Hsp70-1, Hsp70.1, hsp68; heat shock
protein 1B; K03283 heat shock 70kDa protein 1/8
Length=642
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 49/67 (73%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A DKSTGK +ITI + G LS+ +I++MV++AER+K ED+ Q+D VAAKN E
Sbjct 486 LNVT--ATDKSTGKANKITITNDKGRLSKEEIERMVQEAERYKAEDEVQRDRVAAKNALE 543
Query 74 TLVYSVE 80
+ ++++
Sbjct 544 SYAFNMK 550
> mmu:193740 Hspa1a, Hsp70-3, Hsp70.3, Hsp72, MGC189852, hsp68,
hsp70A1; heat shock protein 1A; K03283 heat shock 70kDa protein
1/8
Length=641
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/67 (49%), Positives = 49/67 (73%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A DKSTGK +ITI + G LS+ +I++MV++AER+K ED+ Q+D VAAKN E
Sbjct 486 LNVT--ATDKSTGKANKITITNDKGRLSKEEIERMVQEAERYKAEDEVQRDRVAAKNALE 543
Query 74 TLVYSVE 80
+ ++++
Sbjct 544 SYAFNMK 550
> hsa:3312 HSPA8, HSC54, HSC70, HSC71, HSP71, HSP73, HSPA10, LAP1,
MGC131511, MGC29929, NIP71; heat shock 70kDa protein 8;
K03283 heat shock 70kDa protein 1/8
Length=646
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 47/61 (77%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV++AE++K ED++Q+D V++KN E+ +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSLESYAFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> xla:100037033 hspa1b; heat shock 70kDa protein 1B; K03283 heat
shock 70kDa protein 1/8
Length=646
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 47/61 (77%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV++AE++K ED++Q+D V++KN E+ +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSLESYAFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> mmu:15481 Hspa8, 2410008N15Rik, Hsc70, Hsc71, Hsc73, Hsp73,
Hspa10, MGC102007, MGC106514, MGC118485; heat shock protein
8; K03283 heat shock 70kDa protein 1/8
Length=646
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 47/61 (77%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV++AE++K ED++Q+D V++KN E+ +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDEKQRDKVSSKNSLESYAFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> bbo:BBOV_III010010 17.m07869; dnaK protein; K03283 heat shock
70kDa protein 1/8
Length=647
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 46/66 (69%), Gaps = 3/66 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A+DKSTGK + +TI + G LS A I++MV +AE+FK+ED+ ++ V AK++ E
Sbjct 487 LNVT--AMDKSTGKSEHVTITNDKGRLSTADIERMVAEAEKFKEEDEARRSCVEAKHQLE 544
Query 74 TLVYSV 79
YS+
Sbjct 545 NYCYSM 550
> dre:798846 novel protein similar to vertebrate heat shock 70kDa
protein 1B (HSPA1B); K03283 heat shock 70kDa protein 1/8
Length=639
Score = 55.1 bits (131), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 45/61 (73%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
A DKSTGK +ITI + G LS+ I++MV+DAE++K ED+ Q++ +AAKN E+ +S+
Sbjct 490 AADKSTGKENKITITNDKGRLSKEDIERMVQDAEKYKAEDEVQREKIAAKNALESYAFSM 549
Query 80 E 80
+
Sbjct 550 K 550
> mmu:15482 Hspa1l, Hsc70t, MGC150263, MGC150264, Msh5; heat shock
protein 1-like; K03283 heat shock 70kDa protein 1/8
Length=641
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 50/67 (74%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A+DKSTGK +ITI + G LS+ +I++MV++AER+K ED+ Q++ +AAKN E
Sbjct 488 LNVT--AMDKSTGKANKITITNDKGRLSKEEIERMVQEAERYKAEDEGQREKIAAKNALE 545
Query 74 TLVYSVE 80
+ ++++
Sbjct 546 SYAFNMK 552
> hsa:3310 HSPA6; heat shock 70kDa protein 6 (HSP70B'); K03283
heat shock 70kDa protein 1/8
Length=643
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 44/61 (72%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
A D+STGK +ITI + G LS+ ++++MV +AE++K ED+ Q+D VAAKN E V+ V
Sbjct 492 ATDRSTGKANKITITNDKGRLSKEEVERMVHEAEQYKAEDEAQRDRVAAKNSLEAHVFHV 551
Query 80 E 80
+
Sbjct 552 K 552
> xla:379760 hspa1l, MGC52655, hsc70, hsc70.I; heat shock 70kDa
protein 1-like; K03283 heat shock 70kDa protein 1/8
Length=650
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 47/61 (77%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV++A+++K ED++Q+D V++KN E+ +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKEDIERMVQEADKYKAEDEKQRDKVSSKNSLESYAFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> tpv:TP02_0148 heat shock protein 70; K03283 heat shock 70kDa
protein 1/8
Length=647
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 49/67 (73%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A+DKSTGK + +TI + G LS+ +I +MVE+AE++K+ED++++ V +K++ E
Sbjct 487 LNVT--AMDKSTGKSEHVTITNDKGRLSQEEIDRMVEEAEKYKEEDEKRRKCVESKHKLE 544
Query 74 TLVYSVE 80
YS++
Sbjct 545 NYCYSMK 551
> hsa:3305 HSPA1L, HSP70-1L, HSP70-HOM, HSP70T, hum70t; heat shock
70kDa protein 1-like; K03283 heat shock 70kDa protein 1/8
Length=641
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 48/67 (71%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A DKSTGK +ITI + G LS+ +I++MV DAE++K ED+ Q++ +AAKN E
Sbjct 488 LNVT--ATDKSTGKVNKITITNDKGRLSKEEIERMVLDAEKYKAEDEVQREKIAAKNALE 545
Query 74 TLVYSVE 80
+ ++++
Sbjct 546 SYAFNMK 552
> dre:573376 hspa8, MGC55272, hsc70, wu:fb01g06, wu:fi48b06; heat
shock protein 8; K03283 heat shock 70kDa protein 1/8
Length=649
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/61 (47%), Positives = 45/61 (73%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV++AE++K ED Q+D V+AKN E+ +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDDVQRDKVSAKNGLESYAFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> xla:444468 hspa2, MGC81782; heat shock 70kDa protein 2; K03283
heat shock 70kDa protein 1/8
Length=634
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/67 (47%), Positives = 45/67 (67%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A DKSTGK +ITI + G LS+ I++MV DA+++K ED+ ++ VAAKN E
Sbjct 489 LNVT--AADKSTGKENKITITNDKGRLSKEDIERMVNDADKYKAEDEVNRERVAAKNGLE 546
Query 74 TLVYSVE 80
+ Y V+
Sbjct 547 SYTYHVK 553
> xla:379319 hspa8, MGC53952, hsc54, hsc70, hsc71, hsp71, hsp73,
hspa10, lap1, nip71; heat shock 70kDa protein 8; K03283 heat
shock 70kDa protein 1/8
Length=646
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 46/61 (75%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV++AE +K ED++Q+D V++KN E+ +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKEDIERMVQEAETYKAEDEQQRDKVSSKNSLESYAFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> eco:b0014 dnaK, ECK0014, groPAB, groPC, groPF, grpC, grpF, JW0013,
seg; chaperone HSP70, co-chaperone with DnaJ; K04043
molecular chaperone DnaK
Length=638
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 46/62 (74%), Gaps = 0/62 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A DK++GK Q+ITI++S GL+E +I++MV DAE + D++ ++LV +N+ + L++S
Sbjct 488 AKDKNSGKEQKITIKASSGLNEDEIQKMVRDAEANAEADRKFEELVQTRNQGDHLLHSTR 547
Query 81 KQ 82
KQ
Sbjct 548 KQ 549
> cpv:cgd2_20 heat shock 70 (HSP70) protein ; K03283 heat shock
70kDa protein 1/8
Length=682
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 28/60 (46%), Positives = 42/60 (70%), Gaps = 1/60 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV DAE++K ED++ + + AKN E +Y++
Sbjct 503 AVDKSTGKSSKITITNDKGRLSKDDIERMVNDAEKYKGEDEQNRLKIEAKNSLENYLYNM 562
> hsa:3303 HSPA1A, FLJ54303, FLJ54370, FLJ54392, FLJ54408, FLJ75127,
HSP70-1, HSP70-1A, HSP70I, HSP72, HSPA1, HSPA1B; heat
shock 70kDa protein 1A; K03283 heat shock 70kDa protein 1/8
Length=641
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/67 (44%), Positives = 49/67 (73%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A DKSTGK +ITI + G LS+ +I++MV++AE++K ED+ Q++ V+AKN E
Sbjct 486 LNVT--ATDKSTGKANKITITNDKGRLSKEEIERMVQEAEKYKAEDEVQRERVSAKNALE 543
Query 74 TLVYSVE 80
+ ++++
Sbjct 544 SYAFNMK 550
> hsa:3304 HSPA1B, FLJ54328, HSP70-1B, HSP70-2, HSPA1A; heat shock
70kDa protein 1B; K03283 heat shock 70kDa protein 1/8
Length=641
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/67 (44%), Positives = 49/67 (73%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T A DKSTGK +ITI + G LS+ +I++MV++AE++K ED+ Q++ V+AKN E
Sbjct 486 LNVT--ATDKSTGKANKITITNDKGRLSKEEIERMVQEAEKYKAEDEVQRERVSAKNALE 543
Query 74 TLVYSVE 80
+ ++++
Sbjct 544 SYAFNMK 550
> cel:F26D10.3 hsp-1; Heat Shock Protein family member (hsp-1);
K03283 heat shock 70kDa protein 1/8
Length=640
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 44/61 (72%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
A DKSTGK+ +ITI + G LS+ I++MV +AE++K +D+ QKD + AKN E+ +++
Sbjct 491 ATDKSTGKQNKITITNDKGRLSKDDIERMVNEAEKYKADDEAQKDRIGAKNGLESYAFNL 550
Query 80 E 80
+
Sbjct 551 K 551
> pfa:PF08_0054 heat shock 70 kDa protein; K03283 heat shock 70kDa
protein 1/8
Length=677
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 30/67 (44%), Positives = 43/67 (64%), Gaps = 3/67 (4%)
Query 15 LNQTDRAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAE 73
LN T AV+KSTGK+ ITI + G LS+ +I +MV DAE++K ED+ + + A+N E
Sbjct 499 LNVT--AVEKSTGKQNHITITNDKGRLSQDEIDRMVNDAEKYKAEDEENRKRIEARNSLE 556
Query 74 TLVYSVE 80
Y V+
Sbjct 557 NYCYGVK 563
> dre:393586 MGC63663; zgc:63663; K03283 heat shock 70kDa protein
1/8
Length=647
Score = 51.2 bits (121), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 46/61 (75%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV++A++++ ED+ Q++ V AKN E+L +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKEDIERMVQEADQYRAEDEVQREKVTAKNTLESLAFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> sce:YJR045C SSC1, ENS1; Hsp70 family ATPase, constituent of
the import motor component of the Translocase of the Inner Mitochondrial
membrane (TIM23 complex); involved in protein translocation
and folding; subunit of SceI endonuclease; K04043
molecular chaperone DnaK
Length=654
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 38/60 (63%), Gaps = 0/60 (0%)
Query 21 AVDKSTGKRQQITIQSSGGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSVE 80
A DK+T K IT+ S GLSE +I+QMV DAE+FK +D+ +K + N+A+ L E
Sbjct 511 ARDKATNKDSSITVAGSSGLSENEIEQMVNDAEKFKSQDEARKQAIETANKADQLANDTE 570
> ath:AT5G28540 BIP1; BIP1; ATP binding; K09490 heat shock 70kDa
protein 5
Length=669
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 47/64 (73%), Gaps = 1/64 (1%)
Query 20 RAVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYS 78
+A DK++GK ++ITI + G LS+ +I +MV++AE F +ED++ K+ + A+N ET VY+
Sbjct 520 KAEDKASGKSEKITITNEKGRLSQEEIDRMVKEAEEFAEEDKKVKEKIDARNALETYVYN 579
Query 79 VEKQ 82
++ Q
Sbjct 580 MKNQ 583
> dre:562935 heat shock cognate 70 kDa protein
Length=647
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 45/61 (73%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
AVDKSTGK +ITI + G LS+ I++MV++AE++K ED Q++ V+AKN E+ +++
Sbjct 490 AVDKSTGKENKITITNDKGRLSKEDIERMVQEAEKYKAEDDVQREKVSAKNGLESYSFNM 549
Query 80 E 80
+
Sbjct 550 K 550
> tgo:TGME49_073760 heat shock protein 70, putative ; K03283 heat
shock 70kDa protein 1/8
Length=674
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/58 (50%), Positives = 39/58 (67%), Gaps = 1/58 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVY 77
A DKSTGK QITI + G LS ++I +MV++AE++K ED++ K V AKN E Y
Sbjct 492 AQDKSTGKSNQITITNDKGRLSASEIDRMVQEAEKYKAEDEQNKHRVEAKNGLENYCY 549
> dre:100126123 zgc:174006
Length=643
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 46/61 (75%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
A DKSTGK+ +ITI + G LS+ +I++MV++A+++K ED Q++ ++AKN E+ +++
Sbjct 492 AADKSTGKQNKITITNDKGRLSKEEIERMVQEADKYKAEDDLQREKISAKNSLESYAFNM 551
Query 80 E 80
+
Sbjct 552 K 552
> dre:560210 hsp70l, hsp70-4; heat shock cognate 70-kd protein,
like; K03283 heat shock 70kDa protein 1/8
Length=643
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 46/61 (75%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
A DKSTGK+ +ITI + G LS+ +I++MV++A+++K ED Q++ ++AKN E+ +++
Sbjct 492 AADKSTGKQNRITITNDKGRLSKEEIERMVQEADKYKAEDDLQREKISAKNSLESYAFNM 551
Query 80 E 80
+
Sbjct 552 K 552
> ath:AT1G56410 ERD2; ERD2 (EARLY-RESPONSIVE TO DEHYDRATION 2);
ATP binding; K03283 heat shock 70kDa protein 1/8
Length=617
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/58 (46%), Positives = 40/58 (68%), Gaps = 1/58 (1%)
Query 23 DKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
DK+TGK+ +ITI + G LS+ I++MV++AE++K ED+ K V AKN E Y+V
Sbjct 498 DKATGKKNKITITNDKGRLSKDDIEKMVQEAEKYKSEDEEHKKKVEAKNGLENYAYNV 555
> dre:378848 hspa5, cb865, fb60h09, fi36d04, wu:fb60h09, wu:fi36d04,
zgc:55994, zgc:77606; heat shock protein 5; K09490 heat
shock 70kDa protein 5
Length=650
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 42/63 (66%), Gaps = 1/63 (1%)
Query 21 AVDKSTGKRQQITIQSS-GGLSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
A DK TG + +ITI + L+ I++MV +AERF DED++ K+ + ++NE E+ YS+
Sbjct 511 AEDKGTGNKNKITITNDQNRLTPEDIERMVNEAERFADEDKKLKERIDSRNELESYAYSL 570
Query 80 EKQ 82
+ Q
Sbjct 571 KNQ 573
> ath:AT5G02490 heat shock cognate 70 kDa protein 2 (HSC70-2)
(HSP70-2); K03283 heat shock 70kDa protein 1/8
Length=653
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 41/60 (68%), Gaps = 1/60 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
A DK+TGK+ +ITI + G LS+ I++MV++AE++K ED+ K V AKN E Y++
Sbjct 496 AEDKTTGKKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNM 555
> dre:30671 hsp70, hsp70-4, hspa1a; heat shock cognate 70-kd protein;
K03283 heat shock 70kDa protein 1/8
Length=658
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 45/61 (73%), Gaps = 1/61 (1%)
Query 21 AVDKSTGKRQQITIQSSGG-LSEAQIKQMVEDAERFKDEDQRQKDLVAAKNEAETLVYSV 79
A DKSTGK+ +ITI + G LS+ +I++MV++A+ +K ED Q++ ++AKN E+ +++
Sbjct 492 AADKSTGKQNKITITNDKGRLSKEEIERMVQEADMYKAEDDLQREKISAKNSLESYAFNM 551
Query 80 E 80
+
Sbjct 552 K 552
Lambda K H
0.305 0.124 0.321
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2049280912
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40