bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5202_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
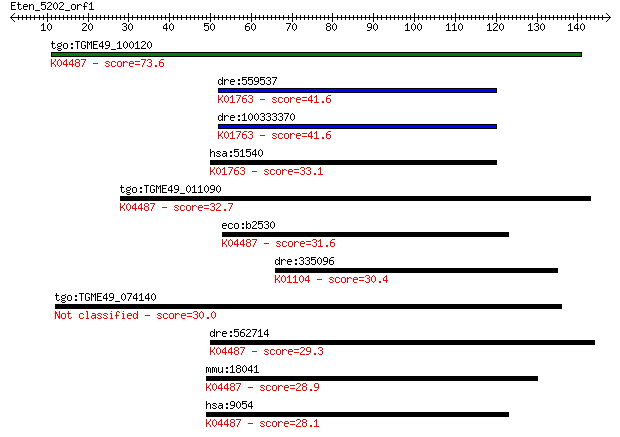
tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);... 73.6 2e-13
dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16... 41.6 0.001
dre:100333370 selenocysteine lyase-like; K01763 selenocysteine... 41.6 0.001
hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);... 33.1 0.35
tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);... 32.7 0.39
eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine de... 31.6 0.84
dre:335096 ptprub, RPTP-psi, fj42a04, fk66g06, ptpru, wu:fj42a... 30.4 1.8
tgo:TGME49_074140 RNA recognition motif-containing protein 30.0 2.6
dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation 1... 29.3 4.1
mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation ge... 28.9 5.3
hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog (S... 28.1 9.0
> tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=851
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 53/144 (36%), Positives = 76/144 (52%), Gaps = 16/144 (11%)
Query 11 FVEAFWATLEQDTKSSRQLLAAAVRFNGPLGAADA------QVQQQW-------LPGTIH 57
F + F+A L T+ S L +R NGPL A A Q++ LP T+
Sbjct 683 FTQQFFAELLLLTQWSPDQLGNLIRINGPLRRARAFFESGDNAQEKSFAEVYGALPNTLS 742
Query 58 ISIWGASGFEIANALSEEERLFVSAGVSCHECKTASGTLLAMGHLD-EWAVGSLRISVGR 116
+SI GA G EI L + RL +SAG +CH + + L LD +WA G++RIS GR
Sbjct 743 LSICGADGPEIVRLLCD--RLCISAGCTCHSSGELTSSTLQAIQLDRKWARGTIRISTGR 800
Query 117 WTSPFEAADAARRVAKLLVSHQLL 140
+T+ +AA A R +A+ L+S +L
Sbjct 801 FTTLNDAAVAGRLLARFLISENML 824
> dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=450
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 29/70 (41%), Positives = 37/70 (52%), Gaps = 3/70 (4%)
Query 52 LPGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK--TASGTLLAMGHLDEWAVGS 109
LP T ++SI G G + LS RL S G +CH + S LL+ G + A +
Sbjct 360 LPNTCNLSILG-RGLQGRRVLSTCRRLLASVGAACHSDRGDQPSHILLSCGIPFDVATNA 418
Query 110 LRISVGRWTS 119
LRISVGR TS
Sbjct 419 LRISVGRSTS 428
> dre:100333370 selenocysteine lyase-like; K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=450
Score = 41.6 bits (96), Expect = 0.001, Method: Composition-based stats.
Identities = 29/70 (41%), Positives = 37/70 (52%), Gaps = 3/70 (4%)
Query 52 LPGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK--TASGTLLAMGHLDEWAVGS 109
LP T ++SI G G + LS RL S G +CH + S LL+ G + A +
Sbjct 360 LPNTCNLSILG-RGLQGRRVLSTCRRLLASVGAACHSDRGDQPSHILLSCGIPFDVATNA 418
Query 110 LRISVGRWTS 119
LRISVGR TS
Sbjct 419 LRISVGRSTS 428
> hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=453
Score = 33.1 bits (74), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 35/72 (48%), Gaps = 3/72 (4%)
Query 50 QWLPGTIHISIWGASGFEIANALSEEERLFVSAGVSCH--ECKTASGTLLAMGHLDEWAV 107
Q LP T + SI G + L++ L S G +CH S LL+ G + A
Sbjct 361 QRLPNTCNFSIRGPR-LQGHVVLAQCRVLMASVGAACHSDHGDQPSPVLLSYGVPFDVAR 419
Query 108 GSLRISVGRWTS 119
+LR+SVGR T+
Sbjct 420 NALRLSVGRSTT 431
> tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=487
Score = 32.7 bits (73), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 30/116 (25%), Positives = 55/116 (47%), Gaps = 7/116 (6%)
Query 28 QLLAAAVRFNGPLGAADAQVQQQWLPGTIHISIWGASGFEIANALSEEERLFVSAGVSCH 87
+LL +VR P + + ++ PG ++IS G + ++ + + +S+G +C
Sbjct 346 RLLLDSVREQIPDIEVNGSLTSRY-PGNLNISFTFVEGESVLMSIRD---VAISSGSACT 401
Query 88 ECK-TASGTLLAMGHLDEWAVGSLRISVGRWTSPFEAADAARRVAKLLVSHQLLQL 142
S L A+G +E A SLR +GR+T + R+ K + H+L +L
Sbjct 402 SASLEPSYVLRALGVGEEVAHTSLRFGIGRFTREEDVRQCVERLVKQI--HRLREL 455
> eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine
desulfurase (tRNA sulfurtransferase), PLP-dependent (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=404
Score = 31.6 bits (70), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 36/71 (50%), Gaps = 4/71 (5%)
Query 53 PGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK-TASGTLLAMGHLDEWAVGSLR 111
P +++S G + AL + L VS+G +C S L A+G DE A S+R
Sbjct 298 PNILNVSFNYVEGESLIMALKD---LAVSSGSACTSASLEPSYVLRALGLNDELAHSSIR 354
Query 112 ISVGRWTSPFE 122
S+GR+T+ E
Sbjct 355 FSLGRFTTEEE 365
> dre:335096 ptprub, RPTP-psi, fj42a04, fk66g06, ptpru, wu:fj42a04,
wu:fk66g06; protein tyrosine phosphatase, receptor type,
U, b (EC:3.1.3.48); K01104 protein-tyrosine phosphatase [EC:3.1.3.48]
Length=1434
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 31/71 (43%), Gaps = 3/71 (4%)
Query 66 FEIANALSEEERLFVSAGVSCHECKTASGTLLAMGHLDEWAVGSLRISVGRWTSPFEAAD 125
F + L E+ER+ +S + H C T S +M GS R GR SP+
Sbjct 800 FTDQSTLQEDERMALSF-MDTHTCSTRSDPRSSMNESSSLLGGSPRRQCGRKGSPYHTGQ 858
Query 126 --AARRVAKLL 134
A RVA LL
Sbjct 859 LHPAVRVADLL 869
> tgo:TGME49_074140 RNA recognition motif-containing protein
Length=788
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 32/126 (25%), Positives = 48/126 (38%), Gaps = 17/126 (13%)
Query 12 VEAFWATLEQDTKSSRQLLAAAVRFNGPLGAADAQVQQQWLPGTIHISIWGASGFEIANA 71
+ A WA + K S + V G +GA W PGT + + +
Sbjct 610 IRATWAKKDSLAKHSSTTDISCVTGTGSIGAEYFGDTGNWQPGTPFVPV---QSHQTMPP 666
Query 72 LSEEERLF--VSAGVSCHECKTASGTLLAMGHLDEWAVGSLRISVGRWTSPFEAADAARR 129
L EE R VS GV + MG+L++ ++ VG +P A D A R
Sbjct 667 LPEEPRRINGVSPGVPTRQ-------PPGMGNLEQ-----RKVPVGACVTPQSAQDFACR 714
Query 130 VAKLLV 135
V + +
Sbjct 715 VCRKFI 720
> dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation
1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=451
Score = 29.3 bits (64), Expect = 4.1, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 45/95 (47%), Gaps = 9/95 (9%)
Query 50 QWLPGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK-TASGTLLAMGHLDEWAVG 108
Q PG I++S G + AL + + +S+G +C S L A+G ++ A
Sbjct 342 QRYPGCINLSFAYVEGESLLMALKD---VALSSGSACTSASLEPSYVLRAIGADEDLAHS 398
Query 109 SLRISVGRWTSPFEAADAARRVAKLLVSHQLLQLK 143
S+R +GR+T+ E A + HQ+ +L+
Sbjct 399 SIRFGIGRFTTEEEVDYTAEKCI-----HQVKRLR 428
> mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation
gene 1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=459
Score = 28.9 bits (63), Expect = 5.3, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 40/82 (48%), Gaps = 4/82 (4%)
Query 49 QQWLPGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK-TASGTLLAMGHLDEWAV 107
+Q PG I++S G + AL + + +S+G +C S L A+G ++ A
Sbjct 349 KQHYPGCINLSFAYVEGESLLMALKD---VALSSGSACTSASLEPSYVLRAIGTDEDLAH 405
Query 108 GSLRISVGRWTSPFEAADAARR 129
S+R +GR+T+ E A +
Sbjct 406 SSIRFGIGRFTTEEEVDYTAEK 427
> hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog
(S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=406
Score = 28.1 bits (61), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 37/75 (49%), Gaps = 4/75 (5%)
Query 49 QQWLPGTIHISIWGASGFEIANALSEEERLFVSAGVSCHECK-TASGTLLAMGHLDEWAV 107
+ PG I++S G + AL + + +S+G +C S L A+G ++ A
Sbjct 296 KHHYPGCINLSFAYVEGESLLMALKD---VALSSGSACTSASLEPSYVLRAIGTDEDLAH 352
Query 108 GSLRISVGRWTSPFE 122
S+R +GR+T+ E
Sbjct 353 SSIRFGIGRFTTEEE 367
Lambda K H
0.319 0.130 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40