bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5144_orf1
Length=109
Score E
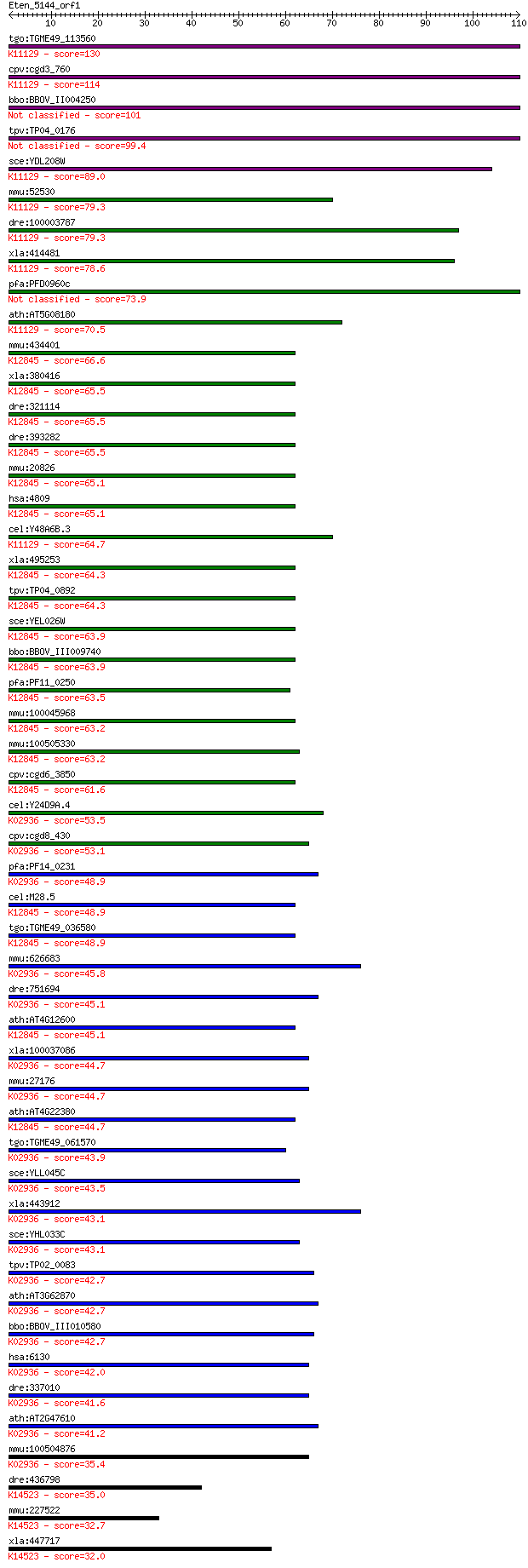
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_113560 60S ribosomal protein L7a, putative ; K11129... 130 1e-30
cpv:cgd3_760 HMG-like nuclear protein, Nhp2p, pelota RNA bindi... 114 5e-26
bbo:BBOV_II004250 18.m06353; ribosomal protein L7Ae-related pr... 101 5e-22
tpv:TP04_0176 40S ribosomal protein L7Ae 99.4 2e-21
sce:YDL208W NHP2; Nhp2p; K11129 H/ACA ribonucleoprotein comple... 89.0 3e-18
mmu:52530 Nhp2, 2410130M07Rik, D11Ertd175e, Nola2; NHP2 ribonu... 79.3 3e-15
dre:100003787 nhp2, MGC73227, nola2, wu:fb36b10, wu:fd61a03, z... 79.3 3e-15
xla:414481 nhp2, MGC81502; NHP2 ribonucleoprotein homolog; K11... 78.6 4e-15
pfa:PFD0960c 60S ribosomal protein L7Ae/L30e, putative 73.9 1e-13
ath:AT5G08180 ribosomal protein L7Ae/L30e/S12e/Gadd45 family p... 70.5 1e-12
mmu:434401 Gm5616, EG434401; predicted gene 5616; K12845 U4/U6... 66.6 2e-11
xla:380416 nhp2l1-b, MGC53430, fa-1, fa1, hoip, nhpx, otk27, s... 65.5 4e-11
dre:321114 nhp2l1, Ssfa1, cb127, sb:cb127, zgc:56265; NHP2 non... 65.5 4e-11
dre:393282 MGC56066, MGC77417, zgc:77417; zgc:56066; K12845 U4... 65.5 4e-11
mmu:20826 Nhp2l1, FA-1, Fta1, Ssfa1; NHP2 non-histone chromoso... 65.1 6e-11
hsa:4809 NHP2L1, 15.5K, FA-1, FA1, NHPX, OTK27, SNRNP15-5, SNU... 65.1 6e-11
cel:Y48A6B.3 hypothetical protein; K11129 H/ACA ribonucleoprot... 64.7 7e-11
xla:495253 nhp2l1-a, fa-1, fa1, hoip, nhp2l1, nhpx, otk27, snr... 64.3 8e-11
tpv:TP04_0892 hypothetical protein; K12845 U4/U6 small nuclear... 64.3 8e-11
sce:YEL026W SNU13; RNA binding protein, part of U3 snoRNP invo... 63.9 1e-10
bbo:BBOV_III009740 17.m07845; ribosomal protein L7A; K12845 U4... 63.9 1e-10
pfa:PF11_0250 high mobility group-like protein NHP2, putative;... 63.5 1e-10
mmu:100045968 NHP2-like protein 1-like; K12845 U4/U6 small nuc... 63.2 2e-10
mmu:100505330 NHP2-like protein 1-like; K12845 U4/U6 small nuc... 63.2 2e-10
cpv:cgd6_3850 HOI-POLLOI protein; U4/U6.U5 snRNP component; Sn... 61.6 6e-10
cel:Y24D9A.4 rpl-7A; Ribosomal Protein, Large subunit family m... 53.5 2e-07
cpv:cgd8_430 60S ribosomal protein L7A ; K02936 large subunit ... 53.1 2e-07
pfa:PF14_0231 60S ribosomal protein L7-3, putative; K02936 lar... 48.9 4e-06
cel:M28.5 hypothetical protein; K12845 U4/U6 small nuclear rib... 48.9 4e-06
tgo:TGME49_036580 60S ribosomal protein L7a, putative ; K12845... 48.9 4e-06
mmu:626683 Gm11362, OTTMUSG00000000657; predicted gene 11362; ... 45.8 3e-05
dre:751694 MGC153325; zgc:153325; K02936 large subunit ribosom... 45.1 5e-05
ath:AT4G12600 ribosomal protein L7Ae/L30e/S12e/Gadd45 family p... 45.1 6e-05
xla:100037086 hypothetical protein LOC100037086; K02936 large ... 44.7 7e-05
mmu:27176 Rpl7a, MGC103146, Surf3; ribosomal protein L7A; K029... 44.7 7e-05
ath:AT4G22380 ribosomal protein L7Ae/L30e/S12e/Gadd45 family p... 44.7 7e-05
tgo:TGME49_061570 60S ribosomal protein L7a, putative ; K02936... 43.9 1e-04
sce:YLL045C RPL8B, KRB1, SCL41; Rpl8bp; K02936 large subunit r... 43.5 2e-04
xla:443912 rpl7a, MGC80199, surf3, trup; ribosomal protein L7a... 43.1 2e-04
sce:YHL033C RPL8A, MAK7; Rpl8ap; K02936 large subunit ribosoma... 43.1 2e-04
tpv:TP02_0083 60S ribosomal protein L7a; K02936 large subunit ... 42.7 3e-04
ath:AT3G62870 60S ribosomal protein L7A (RPL7aB); K02936 large... 42.7 3e-04
bbo:BBOV_III010580 17.m07914; ribosomal protein L7Ae family pr... 42.7 3e-04
hsa:6130 RPL7A, SURF3, TRUP; ribosomal protein L7a; K02936 lar... 42.0 5e-04
dre:337010 rpl7a, wu:fa95b03, zgc:73183, zgc:86667; ribosomal ... 41.6 6e-04
ath:AT2G47610 60S ribosomal protein L7A (RPL7aA); K02936 large... 41.2 9e-04
mmu:100504876 60S ribosomal protein L7a-like; K02936 large sub... 35.4 0.041
dre:436798 rpp38, fc10d08, wu:fc10d08, zgc:92924; ribonuclease... 35.0 0.058
mmu:227522 Rpp38, C330006A15Rik; ribonuclease P/MRP 38 subunit... 32.7 0.26
xla:447717 rpp38, MGC81745; ribonuclease P/MRP 38kDa subunit; ... 32.0 0.45
> tgo:TGME49_113560 60S ribosomal protein L7a, putative ; K11129
H/ACA ribonucleoprotein complex subunit 2
Length=173
Score = 130 bits (327), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 64/110 (58%), Positives = 78/110 (70%), Gaps = 8/110 (7%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GVHEVTK LRKG G+VF ASDV+P+EI AH+PILCE+K++ YAYL KKTLGHAF SKR
Sbjct 71 GVHEVTKCLRKGVKGIVFFASDVFPIEIIAHLPILCEEKDVVYAYLCSKKTLGHAFRSKR 130
Query 61 PASVIMLAPPPPNSGKNRDDDETN-EAEEEDLQEAYSKLEKTIRKNHPYL 109
PASVIM+ P D E + E EE +E Y K+ K +RK++PY
Sbjct 131 PASVIMITP-------GEDMPEVDGEDSEEKFEEVYKKVAKLVRKSNPYF 173
> cpv:cgd3_760 HMG-like nuclear protein, Nhp2p, pelota RNA binding
domain containing protein ; K11129 H/ACA ribonucleoprotein
complex subunit 2
Length=172
Score = 114 bits (286), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 57/109 (52%), Positives = 74/109 (67%), Gaps = 11/109 (10%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GVHEVTKS+RKG++GLV IA D++PV+I AHIPILCE+KNI Y YLG KKTLG SKR
Sbjct 75 GVHEVTKSIRKGQTGLVLIACDIHPVDIIAHIPILCEEKNIYYGYLGSKKTLGTICKSKR 134
Query 61 PASVIMLAPPPPNSGKNRDDDETNEAEEEDLQEAYSKLEKTIRKNHPYL 109
PASV+M++ +S +++ YSK+ I+K HPY+
Sbjct 135 PASVLMISFNSESS-----------VQDKPFYSIYSKVISNIKKVHPYI 172
> bbo:BBOV_II004250 18.m06353; ribosomal protein L7Ae-related
protein
Length=177
Score = 101 bits (252), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 43/109 (39%), Positives = 68/109 (62%), Gaps = 0/109 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV +VTK++RKG G+VFIA DV+P+++ +H+P++CE+ N+AYAY+ K+ + SKR
Sbjct 69 GVQDVTKAIRKGMDGIVFIACDVHPIDVVSHLPVMCEEANMAYAYVDSKRVISSVCQSKR 128
Query 61 PASVIMLAPPPPNSGKNRDDDETNEAEEEDLQEAYSKLEKTIRKNHPYL 109
P V+++ PP + K ++ + E Y KL IR HP+L
Sbjct 129 PTCVVLIVKPPHDLDKRLQTLVQDKHDRLSYLELYDKLNSAIRDLHPFL 177
> tpv:TP04_0176 40S ribosomal protein L7Ae
Length=177
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 47/109 (43%), Positives = 71/109 (65%), Gaps = 1/109 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV +VTKSLRKG +G+V +ASDV+PV++ +H+P+LCE+ +I+YAY+ K+ L +SKR
Sbjct 70 GVLDVTKSLRKGLTGIVLLASDVHPVDVVSHVPVLCEELSISYAYVASKRVLSDVCHSKR 129
Query 61 PASVIMLAPPPPNSGKNRDDDETNEAEEEDLQEAYSKLEKTIRKNHPYL 109
P +++ P + NR + D E Y K++ IRK+HPYL
Sbjct 130 PTCAVLVVKPLKDFS-NRLKKLPEYDNKLDYTELYHKVDSNIRKHHPYL 177
> sce:YDL208W NHP2; Nhp2p; K11129 H/ACA ribonucleoprotein complex
subunit 2
Length=156
Score = 89.0 bits (219), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 68/103 (66%), Gaps = 7/103 (6%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV EV K+LRKGE GLV IA D+ P ++ +HIP+LCED ++ Y ++ K+ LG A +KR
Sbjct 59 GVKEVVKALRKGEKGLVVIAGDISPADVISHIPVLCEDHSVPYIFIPSKQDLGAAGATKR 118
Query 61 PASVIMLAPPPPNSGKNRDDDETNEAEEEDLQEAYSKLEKTIR 103
P SV+ + P G N+ D N +EE+ +E+++++ K ++
Sbjct 119 PTSVVFIVP-----GSNKKKDGKN--KEEEYKESFNEVVKEVQ 154
> mmu:52530 Nhp2, 2410130M07Rik, D11Ertd175e, Nola2; NHP2 ribonucleoprotein
homolog (yeast); K11129 H/ACA ribonucleoprotein
complex subunit 2
Length=153
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 46/69 (66%), Gaps = 0/69 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV EV K + KGE G++ +A D P+E+ H+P+LCED+N+ Y Y+ K LG A SKR
Sbjct 63 GVKEVQKFVNKGEKGIMVLAGDTLPIEVYCHLPVLCEDQNLPYVYIPSKTDLGAATGSKR 122
Query 61 PASVIMLAP 69
P VIM+ P
Sbjct 123 PTCVIMVKP 131
> dre:100003787 nhp2, MGC73227, nola2, wu:fb36b10, wu:fd61a03,
zgc:73227; NHP2 ribonucleoprotein homolog (yeast); K11129 H/ACA
ribonucleoprotein complex subunit 2
Length=150
Score = 79.3 bits (194), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 40/96 (41%), Positives = 57/96 (59%), Gaps = 10/96 (10%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV EV K + KGE+G+V A D P+++ H+PI+CED+++ YAY+ K LG + SKR
Sbjct 60 GVKEVQKFINKGETGIVVFAGDTLPIDVYCHLPIMCEDRSLPYAYVPSKVDLGSSAGSKR 119
Query 61 PASVIMLAPPPPNSGKNRDDDETNEAEEEDLQEAYS 96
P VIM+ P DE EA +E ++E S
Sbjct 120 PTCVIMIKP----------HDEYKEAYDECVEEVTS 145
> xla:414481 nhp2, MGC81502; NHP2 ribonucleoprotein homolog; K11129
H/ACA ribonucleoprotein complex subunit 2
Length=149
Score = 78.6 bits (192), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/95 (42%), Positives = 52/95 (54%), Gaps = 18/95 (18%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV EV K + KGE G+V +A D P+E+ HIP++CED+ I Y+Y+ K LG A SKR
Sbjct 59 GVKEVQKFINKGEKGIVVMAGDTLPIEVYCHIPVMCEDRGIPYSYVPSKSDLGAAAGSKR 118
Query 61 PASVIMLAPPPPNSGKNRDDDETNEAEEEDLQEAY 95
P VI++ P ED QEAY
Sbjct 119 PTCVILIKP------------------HEDYQEAY 135
> pfa:PFD0960c 60S ribosomal protein L7Ae/L30e, putative
Length=236
Score = 73.9 bits (180), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 59/111 (53%), Gaps = 2/111 (1%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G+ +V K +RKG SG+V IA DV+P+ I H+P CE I Y ++ K L H KR
Sbjct 124 GLTQVIKCIRKGYSGIVLIAIDVFPINIICHLPSFCEQYKIPYTFVTTKNKLAHLCKLKR 183
Query 61 PASVIMLAPPPPN--SGKNRDDDETNEAEEEDLQEAYSKLEKTIRKNHPYL 109
+ + + P + + + + + + + + Q+ Y KL ++KNHP+
Sbjct 184 SITCLFILKPDIHILTFEQTINQHSPKKKIHNYQKLYDKLISGVKKNHPFF 234
> ath:AT5G08180 ribosomal protein L7Ae/L30e/S12e/Gadd45 family
protein; K11129 H/ACA ribonucleoprotein complex subunit 2
Length=156
Score = 70.5 bits (171), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 45/71 (63%), Gaps = 0/71 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV EV KS+R+G+ GL IA ++ P+++ H+PILCE+ + Y Y+ K+ L A +KR
Sbjct 54 GVKEVVKSIRRGQKGLCVIAGNISPIDVITHLPILCEEAGVPYVYVPSKEDLAQAGATKR 113
Query 61 PASVIMLAPPP 71
P +++ P
Sbjct 114 PTCCVLVMLKP 124
> mmu:434401 Gm5616, EG434401; predicted gene 5616; K12845 U4/U6
small nuclear ribonucleoprotein SNU13
Length=128
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 41/61 (67%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S L+ +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GANEATKTLNRGISELIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> xla:380416 nhp2l1-b, MGC53430, fa-1, fa1, hoip, nhpx, otk27,
snrnp15-5, snu13, spag12, ssfa1; NHP2 non-histone chromosome
protein 2-like 1; K12845 U4/U6 small nuclear ribonucleoprotein
SNU13
Length=128
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S + +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GANEATKTLNRGISEFIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> dre:321114 nhp2l1, Ssfa1, cb127, sb:cb127, zgc:56265; NHP2 non-histone
chromosome protein 2-like 1 (S. cerevisiae); K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=128
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S + +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GANEATKTLNRGISEFIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> dre:393282 MGC56066, MGC77417, zgc:77417; zgc:56066; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=128
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S + +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GANEATKTLNRGISEFIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> mmu:20826 Nhp2l1, FA-1, Fta1, Ssfa1; NHP2 non-histone chromosome
protein 2-like 1 (S. cerevisiae); K12845 U4/U6 small nuclear
ribonucleoprotein SNU13
Length=128
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S + +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GANEATKTLNRGISEFIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> hsa:4809 NHP2L1, 15.5K, FA-1, FA1, NHPX, OTK27, SNRNP15-5, SNU13,
SPAG12, SSFA1; NHP2 non-histone chromosome protein 2-like
1 (S. cerevisiae); K12845 U4/U6 small nuclear ribonucleoprotein
SNU13
Length=128
Score = 65.1 bits (157), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S + +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GANEATKTLNRGISEFIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> cel:Y48A6B.3 hypothetical protein; K11129 H/ACA ribonucleoprotein
complex subunit 2
Length=163
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 45/69 (65%), Gaps = 0/69 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G+ +V K LR+ E G+ +A +V P+++ +HIP +CE+K I Y Y+ ++ LG A +R
Sbjct 72 GIKDVQKELRRNEKGICILAGNVSPIDVYSHIPGICEEKEIPYVYIPSREQLGLAVGHRR 131
Query 61 PASVIMLAP 69
P+ +I + P
Sbjct 132 PSILIFVKP 140
> xla:495253 nhp2l1-a, fa-1, fa1, hoip, nhp2l1, nhpx, otk27, snrnp15-5,
snu13, spag12, ssfa1; NHP2 non-histone chromosome
protein 2-like 1; K12845 U4/U6 small nuclear ribonucleoprotein
SNU13
Length=128
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G + + +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GANEATKTLNRGIAEFIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> tpv:TP04_0892 hypothetical protein; K12845 U4/U6 small nuclear
ribonucleoprotein SNU13
Length=129
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G + +V +A+D P+EI H+P+LCEDKN+ Y ++ K LG A R
Sbjct 39 GANEATKALNRGHAEIVLLAADAEPLEIILHLPLLCEDKNVPYIFVHSKVALGRACGVSR 98
Query 61 P 61
P
Sbjct 99 P 99
> sce:YEL026W SNU13; RNA binding protein, part of U3 snoRNP involved
in rRNA processing, part of U4/U6-U5 tri-snRNP involved
in mRNA splicing, similar to human 15.5K protein; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=126
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 39/61 (63%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S + +A+D P+EI H+P+LCEDKN+ Y ++ + LG A R
Sbjct 36 GANEATKTLNRGISEFIIMAADCEPIEILLHLPLLCEDKNVPYVFVPSRVALGRACGVSR 95
Query 61 P 61
P
Sbjct 96 P 96
> bbo:BBOV_III009740 17.m07845; ribosomal protein L7A; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=128
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TKSL +G + +V +A+D P+EI H+P++CEDKNI Y ++ K LG A R
Sbjct 38 GANEATKSLNRGLAEIVVLAADAEPLEIILHLPLVCEDKNIPYIFVKSKIALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> pfa:PF11_0250 high mobility group-like protein NHP2, putative;
K12845 U4/U6 small nuclear ribonucleoprotein SNU13
Length=145
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 38/60 (63%), Gaps = 0/60 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E KSL KG S LV +A+D P+EI +HIP++CEDKN Y Y+ K LG A R
Sbjct 55 GANEAVKSLHKGISELVVLAADAKPLEIISHIPLVCEDKNTPYVYVRSKMALGRACGISR 114
> mmu:100045968 NHP2-like protein 1-like; K12845 U4/U6 small nuclear
ribonucleoprotein SNU13
Length=134
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+E TK+ +G S + +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GVNEATKTPNRGISEFIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 P 61
P
Sbjct 98 P 98
> mmu:100505330 NHP2-like protein 1-like; K12845 U4/U6 small nuclear
ribonucleoprotein SNU13
Length=128
Score = 63.2 bits (152), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+E TK+ +G S + +A+D P+EI H+P+LCEDKN+ Y ++ K+ LG A R
Sbjct 38 GVNEATKTPNRGISEFIVMAADAEPLEIILHLPLLCEDKNVPYVFVRSKQALGRACGVSR 97
Query 61 PA 62
P
Sbjct 98 PV 99
> cpv:cgd6_3850 HOI-POLLOI protein; U4/U6.U5 snRNP component;
Snu13p; pelota RNA binding domain containing protein ; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=134
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 39/61 (63%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G + +V +A+D P+EI H+P++CEDKN Y ++ K LG A R
Sbjct 44 GANEATKALNRGIAEIVLLAADAEPLEILLHLPLVCEDKNTPYVFVRSKVALGRACGVSR 103
Query 61 P 61
P
Sbjct 104 P 104
> cel:Y24D9A.4 rpl-7A; Ribosomal Protein, Large subunit family
member (rpl-7A); K02936 large subunit ribosomal protein L7Ae
Length=245
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 38/67 (56%), Gaps = 0/67 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+ +T+ + + LV IA DV P+EI H+P LC N+ YA + K +LG K
Sbjct 120 GVNTITRLVETRRAQLVLIAHDVNPLEIVLHLPALCRKYNVPYAIIKGKASLGTVVRRKT 179
Query 61 PASVIML 67
A+V ++
Sbjct 180 TAAVALV 186
> cpv:cgd8_430 60S ribosomal protein L7A ; K02936 large subunit
ribosomal protein L7Ae
Length=263
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 36/64 (56%), Gaps = 0/64 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+ VT + ++ LV IASDV P+EI +P LC K++A+ +G K LG K
Sbjct 134 GVNHVTDLVEMKKAKLVVIASDVDPIEIVCFLPALCRRKDVAFCIIGGKARLGKLIGKKT 193
Query 61 PASV 64
A V
Sbjct 194 AAVV 197
> pfa:PF14_0231 60S ribosomal protein L7-3, putative; K02936 large
subunit ribosomal protein L7Ae
Length=283
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 37/66 (56%), Gaps = 0/66 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G++ +TK + ++ LV IA+DV P+E+ +P LC K + Y + K TLG + K
Sbjct 152 GINHITKLVENKKANLVVIANDVSPIELVLFLPALCRLKEVPYCIVKDKATLGKLVHKKT 211
Query 61 PASVIM 66
+V +
Sbjct 212 ATAVCL 217
> cel:M28.5 hypothetical protein; K12845 U4/U6 small nuclear ribonucleoprotein
SNU13
Length=128
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S ++ +A+D P+EI H+P+LCEDKN+ Y ++ K LG A R
Sbjct 38 GANEATKTLNRGISEIIVMAADAEPLEILLHLPLLCEDKNVPYVFVRSKAALGRACGVTR 97
Query 61 P 61
P
Sbjct 98 P 98
> tgo:TGME49_036580 60S ribosomal protein L7a, putative ; K12845
U4/U6 small nuclear ribonucleoprotein SNU13
Length=131
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G + +V +A+D P+EI H+P+LCEDKN+ Y ++ K LG A R
Sbjct 41 GANEATKTLNRGTAEIVILAADAEPLEILLHLPLLCEDKNVPYVFVRSKVALGRACGVSR 100
Query 61 P 61
P
Sbjct 101 P 101
> mmu:626683 Gm11362, OTTMUSG00000000657; predicted gene 11362;
K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 36/75 (48%), Gaps = 0/75 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+ VT + ++ LV IA DV P+E+ +P LC+ + Y + K LGH + K
Sbjct 139 GVNTVTTLVENKKAQLVVIAHDVDPIELVVFLPALCQKMGVPYCIIKGKARLGHLVHRKT 198
Query 61 PASVIMLAPPPPNSG 75
+V N G
Sbjct 199 CTTVAFTQVNSENKG 213
> dre:751694 MGC153325; zgc:153325; K02936 large subunit ribosomal
protein L7Ae
Length=269
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 36/66 (54%), Gaps = 0/66 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G+++VT+ + + ++ LV IA DV PVE+ +P LC + Y + K LG K
Sbjct 141 GINDVTRLVEQKKAQLVVIAHDVNPVELVIFLPSLCRKMGVPYCIVKDKSRLGRLVRRKT 200
Query 61 PASVIM 66
++V +
Sbjct 201 CSAVAL 206
> ath:AT4G12600 ribosomal protein L7Ae/L30e/S12e/Gadd45 family
protein; K12845 U4/U6 small nuclear ribonucleoprotein SNU13
Length=128
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 26/61 (42%), Positives = 40/61 (65%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S + +A+D P+EI H+P+L EDKN+ Y ++ K+ LG A + R
Sbjct 38 GANEATKTLNRGISEFIVMAADTEPLEILLHLPLLAEDKNVPYVFVPSKQALGRACDVTR 97
Query 61 P 61
P
Sbjct 98 P 98
> xla:100037086 hypothetical protein LOC100037086; K02936 large
subunit ribosomal protein L7Ae
Length=266
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+ VT + ++ LV IA DV P+E+ +P LC + Y + K LGH + K
Sbjct 139 GVNTVTTLVENKKAQLVVIAHDVDPIELVVFLPALCRKMGVPYCIIKGKARLGHLVHRKT 198
Query 61 PASV 64
+V
Sbjct 199 CTTV 202
> mmu:27176 Rpl7a, MGC103146, Surf3; ribosomal protein L7A; K02936
large subunit ribosomal protein L7Ae
Length=266
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+ VT + ++ LV IA DV P+E+ +P LC + Y + K LGH + K
Sbjct 139 GVNTVTTLVENKKAQLVVIAHDVDPIELVVFLPALCRKMGVPYCIIKGKARLGHLVHRKT 198
Query 61 PASV 64
+V
Sbjct 199 CTTV 202
> ath:AT4G22380 ribosomal protein L7Ae/L30e/S12e/Gadd45 family
protein; K12845 U4/U6 small nuclear ribonucleoprotein SNU13
Length=128
Score = 44.7 bits (104), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 39/61 (63%), Gaps = 0/61 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G +E TK+L +G S V +A+D P+EI H+P+L EDKN+ Y ++ K+ LG A R
Sbjct 38 GANEATKTLNRGISEFVVMAADAEPLEILLHLPLLAEDKNVPYVFVPSKQALGRACGVTR 97
Query 61 P 61
P
Sbjct 98 P 98
> tgo:TGME49_061570 60S ribosomal protein L7a, putative ; K02936
large subunit ribosomal protein L7Ae
Length=276
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 31/59 (52%), Gaps = 0/59 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSK 59
G++ VT + ++ LV IA DV PVE+ IP LC K + Y + K LG + K
Sbjct 146 GINHVTDLIEIKKAKLVVIAHDVTPVELVCFIPQLCRKKEVPYCIVKGKSRLGQLVHQK 204
> sce:YLL045C RPL8B, KRB1, SCL41; Rpl8bp; K02936 large subunit
ribosomal protein L7Ae
Length=256
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 33/62 (53%), Gaps = 0/62 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G++ V + ++ LV IA+DV P+E+ +P LC+ + YA + K LG N K
Sbjct 135 GLNHVVSLIENKKAKLVLIANDVDPIELVVFLPALCKKMGVPYAIIKGKARLGTLVNQKT 194
Query 61 PA 62
A
Sbjct 195 SA 196
> xla:443912 rpl7a, MGC80199, surf3, trup; ribosomal protein L7a;
K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 35/75 (46%), Gaps = 0/75 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+ VT + ++ LV IA DV P+E+ +P LC + Y + K LG + K
Sbjct 139 GVNTVTTLVENKKAQLVVIAHDVDPIELVVFLPALCRKMGVPYCIVKGKARLGRLVHRKT 198
Query 61 PASVIMLAPPPPNSG 75
SV P + G
Sbjct 199 CTSVAFAQVNPEDKG 213
> sce:YHL033C RPL8A, MAK7; Rpl8ap; K02936 large subunit ribosomal
protein L7Ae
Length=256
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 33/62 (53%), Gaps = 0/62 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G++ V + ++ LV IA+DV P+E+ +P LC+ + YA + K LG N K
Sbjct 135 GLNHVVALIENKKAKLVLIANDVDPIELVVFLPALCKKMGVPYAIVKGKARLGTLVNQKT 194
Query 61 PA 62
A
Sbjct 195 SA 196
> tpv:TP02_0083 60S ribosomal protein L7a; K02936 large subunit
ribosomal protein L7Ae
Length=266
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G++ VT + ++ LV IA DV P+E+ +P LC K + Y + K LG + K
Sbjct 136 GLNHVTNLVEYKKAKLVVIAHDVDPIELVLWLPALCRKKEVPYCIIKSKSRLGKLVHQK- 194
Query 61 PASVI 65
ASV+
Sbjct 195 TASVV 199
> ath:AT3G62870 60S ribosomal protein L7A (RPL7aB); K02936 large
subunit ribosomal protein L7Ae
Length=256
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 35/66 (53%), Gaps = 0/66 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G++ VT + + ++ LV IA DV P+E+ +P LC + Y + K LG + K
Sbjct 129 GLNHVTYLIEQNKAQLVVIAHDVDPIELVVWLPALCRKMEVPYCIVKGKSRLGAVVHQKT 188
Query 61 PASVIM 66
A++ +
Sbjct 189 AAALCL 194
> bbo:BBOV_III010580 17.m07914; ribosomal protein L7Ae family
protein; K02936 large subunit ribosomal protein L7Ae
Length=269
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 1/65 (1%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G++ VT + ++ LV IA DV P+E+ +P LC K + Y + K LG + +R
Sbjct 139 GLNHVTNLVEYKKAKLVVIAHDVDPLELVMWLPTLCRKKEVPYCIIKGKSRLGKLVH-QR 197
Query 61 PASVI 65
ASV+
Sbjct 198 TASVV 202
> hsa:6130 RPL7A, SURF3, TRUP; ribosomal protein L7a; K02936 large
subunit ribosomal protein L7Ae
Length=266
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+ VT + ++ LV IA DV P+E+ +P LC + Y + K LG + K
Sbjct 139 GVNTVTTLVENKKAQLVVIAHDVDPIELVVFLPALCRKMGVPYCIIKGKARLGRLVHRKT 198
Query 61 PASV 64
+V
Sbjct 199 CTTV 202
> dre:337010 rpl7a, wu:fa95b03, zgc:73183, zgc:86667; ribosomal
protein L7a; K02936 large subunit ribosomal protein L7Ae
Length=266
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 32/64 (50%), Gaps = 0/64 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
GV+ VT + ++ LV IA DV P+E+ +P LC + Y + K LG + K
Sbjct 139 GVNTVTTLVESKKAQLVVIAHDVDPIELVVFLPALCRKMGVPYCIVKGKARLGRLVHRKT 198
Query 61 PASV 64
S+
Sbjct 199 CTSI 202
> ath:AT2G47610 60S ribosomal protein L7A (RPL7aA); K02936 large
subunit ribosomal protein L7Ae
Length=257
Score = 41.2 bits (95), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 34/66 (51%), Gaps = 0/66 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNIAYAYLGPKKTLGHAFNSKR 60
G++ VT + + ++ LV IA DV P+E+ +P LC + Y + K LG + K
Sbjct 130 GLNHVTYLIEQNKAQLVVIAHDVDPIELVVWLPALCRKMEVPYCIVKGKSRLGAVVHQKT 189
Query 61 PASVIM 66
+ + +
Sbjct 190 ASCLCL 195
> mmu:100504876 60S ribosomal protein L7a-like; K02936 large subunit
ribosomal protein L7Ae
Length=278
Score = 35.4 bits (80), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 32/65 (49%), Gaps = 1/65 (1%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVE-ITAHIPILCEDKNIAYAYLGPKKTLGHAFNSK 59
GV+ VT + ++ LV IA DV P+E + +P LC + Y + K LG + K
Sbjct 150 GVNTVTILVENKKAQLVVIAHDVDPIEMLVVFLPALCRKMGVPYCIIKGKARLGCLVHRK 209
Query 60 RPASV 64
+V
Sbjct 210 TCTTV 214
> dre:436798 rpp38, fc10d08, wu:fc10d08, zgc:92924; ribonuclease
P/MRP 38 subunit (EC:3.1.26.5); K14523 ribonucleases P/MRP
protein subunit RPP38 [EC:3.1.26.5]
Length=265
Score = 35.0 bits (79), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHIPILCEDKNI 41
G++EVTK L + E LV + + V P +T+H+ L + +++
Sbjct 113 GINEVTKGLERNELSLVLVCNSVTPAHMTSHLIPLSKTRSV 153
> mmu:227522 Rpp38, C330006A15Rik; ribonuclease P/MRP 38 subunit
(human) (EC:3.1.26.5); K14523 ribonucleases P/MRP protein
subunit RPP38 [EC:3.1.26.5]
Length=280
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAHI 32
GV+EVT++L + E LV + V P IT+H+
Sbjct 111 GVNEVTRALERNELLLVLVCKSVKPAIITSHL 142
> xla:447717 rpp38, MGC81745; ribonuclease P/MRP 38kDa subunit;
K14523 ribonucleases P/MRP protein subunit RPP38 [EC:3.1.26.5]
Length=279
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 30/70 (42%), Gaps = 17/70 (24%)
Query 1 GVHEVTKSLRKGESGLVFIASDVYPVEITAH--------------IPILCEDKNIAYAYL 46
G++EVT+ L K E LV + P IT H IP L E NIA A L
Sbjct 114 GINEVTRGLEKNELSLVLVCKSAKPEMITKHLIELSISRETPACQIPRLSE--NIAPA-L 170
Query 47 GPKKTLGHAF 56
G K L F
Sbjct 171 GLKSVLALGF 180
Lambda K H
0.311 0.132 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2072286120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40