bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5135_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
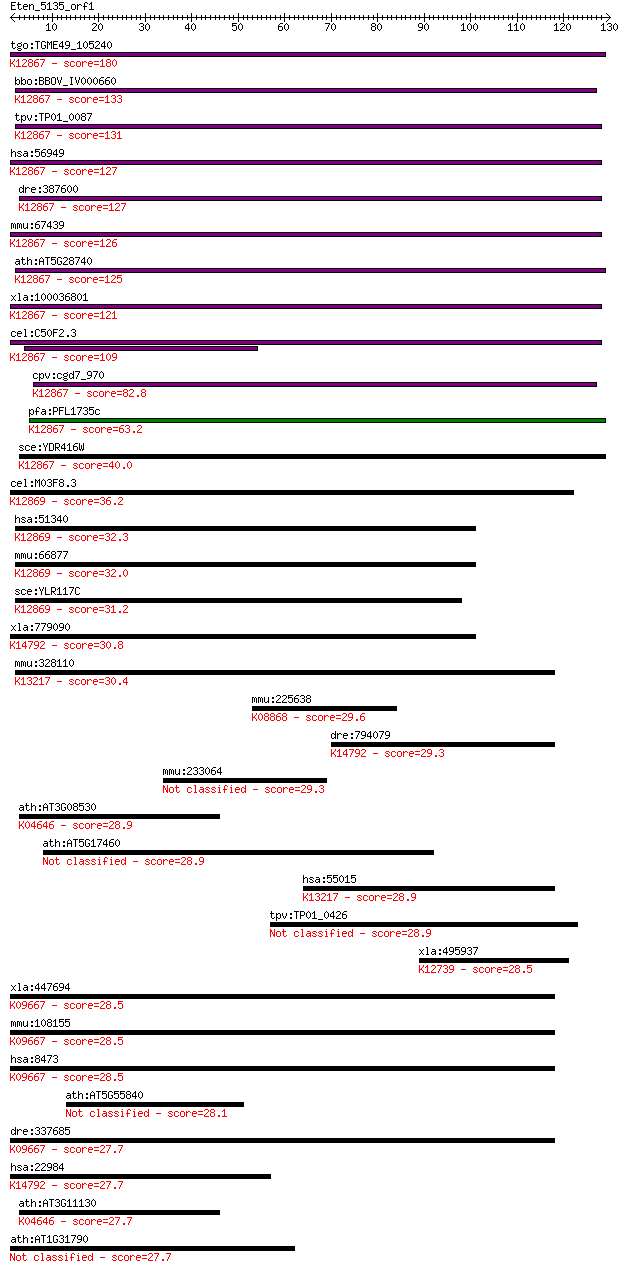
tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-m... 180 1e-45
bbo:BBOV_IV000660 21.m02991; XBA-binding protein 2; K12867 pre... 133 1e-31
tpv:TP01_0087 adapter protein; K12867 pre-mRNA-splicing factor... 131 5e-31
hsa:56949 XAB2, DKFZp762C1015, HCNP, HCRN, NTC90, SYF1; XPA bi... 127 8e-30
dre:387600 xab2, MGC198247, zgc:63498, zgc:63949; XPA binding ... 127 9e-30
mmu:67439 Xab2, 0610041O14Rik, AV025587; XPA binding protein 2... 126 2e-29
ath:AT5G28740 transcription-coupled DNA repair protein-related... 125 5e-29
xla:100036801 xab2; XPA binding protein 2; K12867 pre-mRNA-spl... 121 7e-28
cel:C50F2.3 hypothetical protein; K12867 pre-mRNA-splicing fac... 109 3e-24
cpv:cgd7_970 Syf1p. protein with 8 HAT domains ; K12867 pre-mR... 82.8 3e-16
pfa:PFL1735c RNA-processing protein, putative; K12867 pre-mRNA... 63.2 2e-10
sce:YDR416W SYF1, NTC90; Member of the NineTeen Complex (NTC) ... 40.0 0.002
cel:M03F8.3 hypothetical protein; K12869 crooked neck 36.2 0.024
hsa:51340 CRNKL1, CLF, CRN, Clf1, HCRN, MSTP021, SYF3; crooked... 32.3 0.41
mmu:66877 Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn; C... 32.0 0.45
sce:YLR117C CLF1, NTC77, SYF3; Clf1p; K12869 crooked neck 31.2 0.74
xla:779090 pdcd11, alg-4, alg4, nfbp, rrp5; programmed cell de... 30.8 1.1
mmu:328110 Prpf39, FLJ11128, MGC37077, Srcs1; PRP39 pre-mRNA p... 30.4 1.5
mmu:225638 Alpk2, Gm549, Hak; alpha-kinase 2; K08868 alpha-kinase 29.6 2.7
dre:794079 pdcd11, MGC162501, cb680, im:7148359, sb:cb680, wu:... 29.3 2.9
mmu:233064 Wdr62, 2310038K02Rik, MGC31423; WD repeat domain 62 29.3
ath:AT3G08530 clathrin heavy chain, putative; K04646 clathrin ... 28.9 3.8
ath:AT5G17460 hypothetical protein 28.9 4.0
hsa:55015 PRPF39, FLJ11128, FLJ20666, FLJ45460, MGC149842, MGC... 28.9 4.6
tpv:TP01_0426 hypothetical protein 28.9 4.7
xla:495937 ppil6; peptidylprolyl isomerase (cyclophilin)-like ... 28.5 5.7
xla:447694 ogt, MGC80426; O-linked N-acetylglucosamine (GlcNAc... 28.5 6.2
mmu:108155 Ogt, 1110038P24Rik, 4831420N21Rik, AI115525, Ogtl; ... 28.5 6.2
hsa:8473 OGT, FLJ23071, HRNT1, MGC22921, O-GLCNAC; O-linked N-... 28.5 6.2
ath:AT5G55840 pentatricopeptide (PPR) repeat-containing protein 28.1 7.3
dre:337685 ogt.1, fm81g08, ogt, wu:fc12b01, wu:fm81g08; O-link... 27.7 8.2
hsa:22984 PDCD11, ALG-4, ALG4, KIAA0185, NFBP, RRP5; programme... 27.7 8.3
ath:AT3G11130 clathrin heavy chain, putative; K04646 clathrin ... 27.7 9.6
ath:AT1G31790 pentatricopeptide (PPR) repeat-containing protein 27.7 9.7
> tgo:TGME49_105240 XPA-binding protein, putative ; K12867 pre-mRNA-splicing
factor SYF1
Length=966
Score = 180 bits (456), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 89/128 (69%), Positives = 102/128 (79%), Gaps = 1/128 (0%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQD 60
GDL NA +FE+A +A RT+D+LAS+WCEAVEM LR +K AL LVR AIS R D
Sbjct 458 GDLPNARLIFEKATKARVRTVDELASIWCEAVEMELRREEWKRALELVRRAISRPRDA-D 516
Query 61 TKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLEERR 120
AQ +LFRSVKLWSLAADVEEM GS ETVR CY+KMFQLKVITPQLVINYAH+LEE R
Sbjct 517 PDSAQAKLFRSVKLWSLAADVEEMTGSPETVRLCYNKMFQLKVITPQLVINYAHFLEEHR 576
Query 121 YYEESFRV 128
++EESF+V
Sbjct 577 FFEESFKV 584
> bbo:BBOV_IV000660 21.m02991; XBA-binding protein 2; K12867 pre-mRNA-splicing
factor SYF1
Length=796
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 62/125 (49%), Positives = 88/125 (70%), Gaps = 1/125 (0%)
Query 2 DLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQDT 61
D++ A ++FE+AV+ Y+ +DDLASVWC VEMH+RH K AL L R A+ R ++
Sbjct 453 DVDAADRIFEKAVEGNYKFVDDLASVWCAWVEMHIRHNNLKRALELSRQAVDV-RNKKEP 511
Query 62 KGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLEERRY 121
+ RL+RSVKLWSL D+E+ G+I T RA +D M +LKV+TPQ+ +N+A YLEE +Y
Sbjct 512 NYVEQRLYRSVKLWSLCLDLEQNLGTIATARATFDLMAELKVVTPQIALNFAMYLEEHKY 571
Query 122 YEESF 126
+E +F
Sbjct 572 FEAAF 576
> tpv:TP01_0087 adapter protein; K12867 pre-mRNA-splicing factor
SYF1
Length=839
Score = 131 bits (329), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 61/126 (48%), Positives = 93/126 (73%), Gaps = 4/126 (3%)
Query 2 DLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQDT 61
DLENA K++E+A + ++ +DDLA++WC VEM+LRH FK+AL + R ++S + K +
Sbjct 486 DLENADKIYEKASNSDFKYVDDLATLWCCWVEMYLRHKQFKKALEISRRSVSGNGKTSIS 545
Query 62 KGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLEERRY 121
+ RL SVKLW+LA D+E+ FG+IET RA ++KM +LKV+TPQ+ +++A YLE+ +Y
Sbjct 546 R----RLHSSVKLWTLALDMEQNFGTIETTRATFNKMVELKVVTPQVALSFAGYLEQNKY 601
Query 122 YEESFR 127
+E SF
Sbjct 602 FEASFN 607
> hsa:56949 XAB2, DKFZp762C1015, HCNP, HCRN, NTC90, SYF1; XPA
binding protein 2; K12867 pre-mRNA-splicing factor SYF1
Length=855
Score = 127 bits (319), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 57/131 (43%), Positives = 94/131 (71%), Gaps = 4/131 (3%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAIS-SSRKGQ 59
G L++A + E+A + ++ +DDLASVWC+ E+ LRH + EALRL+R A + +R+ +
Sbjct 409 GQLDDARVILEKATKVNFKQVDDLASVWCQCGELELRHENYDEALRLLRKATALPARRAE 468
Query 60 DTKGA---QGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYL 116
G+ Q R+++S+K+WS+ AD+EE G+ ++ +A YD++ L++ TPQ+VINYA +L
Sbjct 469 YFDGSEPVQNRVYKSLKVWSMLADLEESLGTFQSTKAVYDRILDLRIATPQIVINYAMFL 528
Query 117 EERRYYEESFR 127
EE +Y+EESF+
Sbjct 529 EEHKYFEESFK 539
> dre:387600 xab2, MGC198247, zgc:63498, zgc:63949; XPA binding
protein 2; K12867 pre-mRNA-splicing factor SYF1
Length=851
Score = 127 bits (319), Expect = 9e-30, Method: Compositional matrix adjust.
Identities = 55/129 (42%), Positives = 93/129 (72%), Gaps = 4/129 (3%)
Query 3 LENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAIS-SSRKGQ-- 59
+++A +FE+A + Y+ +DDLA+VWCE EM LRH + +ALR++R A + +RK +
Sbjct 406 IDDARTIFEKATKVNYKQVDDLAAVWCEYGEMELRHENYDQALRILRKATAIPARKAEYF 465
Query 60 -DTKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLEE 118
++ Q R+++S+K+WS+ AD+EE G+ ++ +A YD++ L++ TPQ++INYA +LEE
Sbjct 466 DSSEPVQNRVYKSLKVWSMLADLEESLGTFQSTKAVYDRIIDLRIATPQIIINYAMFLEE 525
Query 119 RRYYEESFR 127
Y+EESF+
Sbjct 526 HNYFEESFK 534
> mmu:67439 Xab2, 0610041O14Rik, AV025587; XPA binding protein
2; K12867 pre-mRNA-splicing factor SYF1
Length=855
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 94/131 (71%), Gaps = 4/131 (3%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAIS-SSRKGQ 59
G L++A + E+A + ++ +DDLASVWC+ E+ LRH + EAL+L+R A + +R+ +
Sbjct 409 GQLDDARVILEKATKVNFKQVDDLASVWCQCGELELRHENYDEALKLLRKATALPARRAE 468
Query 60 DTKGA---QGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYL 116
G+ Q R+++S+K+WS+ AD+EE G+ ++ +A YD++ L++ TPQ+VINYA +L
Sbjct 469 YFDGSEPVQNRVYKSLKVWSMLADLEESLGTFQSTKAVYDRILDLRIATPQIVINYAMFL 528
Query 117 EERRYYEESFR 127
EE +Y+EESF+
Sbjct 529 EEHKYFEESFK 539
> ath:AT5G28740 transcription-coupled DNA repair protein-related;
K12867 pre-mRNA-splicing factor SYF1
Length=917
Score = 125 bits (313), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 61/134 (45%), Positives = 88/134 (65%), Gaps = 7/134 (5%)
Query 2 DLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSS------ 55
DL N +F++AVQ Y+T+D LASVWCE EM LRH FK AL L+R A +
Sbjct 438 DLVNTRVIFDKAVQVNYKTVDHLASVWCEWAEMELRHKNFKGALELMRRATAVPTVEVRR 497
Query 56 RKGQD-TKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAH 114
R D + Q +L RS++LWS D+EE G++E+ RA Y+K+ L++ TPQ+++NYA
Sbjct 498 RVAADGNEPVQMKLHRSLRLWSFYVDLEESLGTLESTRAVYEKILDLRIATPQIIMNYAF 557
Query 115 YLEERRYYEESFRV 128
LEE +Y+E++F+V
Sbjct 558 LLEENKYFEDAFKV 571
> xla:100036801 xab2; XPA binding protein 2; K12867 pre-mRNA-splicing
factor SYF1
Length=838
Score = 121 bits (303), Expect = 7e-28, Method: Compositional matrix adjust.
Identities = 56/131 (42%), Positives = 89/131 (67%), Gaps = 4/131 (3%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAIS-SSRKGQ 59
G +E+A + +RA Y +DDLASVWC+ EM LRH + AL+++R A + +RK +
Sbjct 400 GQIEDARAILQRATLVQYTHVDDLASVWCQFGEMELRHENYDAALKILRKATAVPARKAE 459
Query 60 ---DTKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYL 116
++ Q RL++S+++WS+ AD+EE G+ ++ +A YD++ L + TPQ+VINYA +L
Sbjct 460 YFDSSEPVQNRLYKSLRVWSMLADLEESLGTFKSTKAVYDRIIDLHIATPQIVINYAMFL 519
Query 117 EERRYYEESFR 127
EE Y+EESF+
Sbjct 520 EEHNYFEESFK 530
> cel:C50F2.3 hypothetical protein; K12867 pre-mRNA-splicing factor
SYF1
Length=855
Score = 109 bits (272), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 52/129 (40%), Positives = 83/129 (64%), Gaps = 2/129 (1%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKG-- 58
GDL+ A K FE AV + + + +LA+VWC EM ++H K AL +++ A + G
Sbjct 415 GDLDAARKTFETAVISQFGGVSELANVWCAYAEMEMKHKRAKAALTVMQRACVVPKPGDY 474
Query 59 QDTKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLEE 118
++ + Q R+ RS LW++ AD EE G++E+ R YDKM +L+V +PQ+++NYA +LEE
Sbjct 475 ENMQSVQARVHRSPILWAMYADYEECCGTVESCRKVYDKMIELRVASPQMIMNYAMFLEE 534
Query 119 RRYYEESFR 127
Y+E +F+
Sbjct 535 NEYFELAFQ 543
Score = 27.7 bits (60), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 25/55 (45%), Gaps = 5/55 (9%)
Query 4 ENAAKVFERAV-----QATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAIS 53
+N K+F V Q YR D + +WC + ++R F+ A + AI+
Sbjct 237 KNPVKIFSLNVDAIIRQGIYRYTDQVGFLWCSLADYYIRSAEFERARDVYEEAIA 291
> cpv:cgd7_970 Syf1p. protein with 8 HAT domains ; K12867 pre-mRNA-splicing
factor SYF1
Length=1020
Score = 82.8 bits (203), Expect = 3e-16, Method: Composition-based stats.
Identities = 45/121 (37%), Positives = 75/121 (61%), Gaps = 7/121 (5%)
Query 6 AAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQDTKGAQ 65
A +FER++ Y I+D + +W E +EM LR G F+EAL L R +I +++ Q +K
Sbjct 559 ARDIFERSLSEDY--IEDYSLIWTEWIEMELRFGNFEEALNLSRRSICMAKE-QKSKIT- 614
Query 66 GRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLEERRYYEES 125
R+ ++W+LAAD+E FG++E+ RA + +F+ + T L++ + YL ++ YEES
Sbjct 615 ---LRNGRIWNLAADLEMSFGTLESSRALIEDLFESGMATANLLVTFGSYLRDKECYEES 671
Query 126 F 126
F
Sbjct 672 F 672
> pfa:PFL1735c RNA-processing protein, putative; K12867 pre-mRNA-splicing
factor SYF1
Length=1031
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 42/151 (27%), Positives = 77/151 (50%), Gaps = 27/151 (17%)
Query 5 NAAKVFERAVQ--ATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQDT- 61
N +F+ A++ A +++ +++A+++C +E+ L +KEAL + RL+I ++K +T
Sbjct 632 NCINIFKLALKQNAYFKSANEIANIFCAWIEIELLERNYKEALNIARLSIDINKKSYNTL 691
Query 62 -KGAQG-----------------------RLFRSVKLWSLAADVEEMFGSIETVRACYDK 97
K + L +KL SL D+E +G+IET +D
Sbjct 692 YKSSTSILLYEDISLNNNLKNKNNYHTNFNLLSCMKLVSLIIDMEMNYGTIETTLNMFDF 751
Query 98 MFQLKVITPQLVINYAHYLEERRYYEESFRV 128
+ K I ++V+ A YL E++Y+ ESF+V
Sbjct 752 FYHSKCINVKMVLTLATYLYEKKYFNESFKV 782
> sce:YDR416W SYF1, NTC90; Member of the NineTeen Complex (NTC)
that contains Prp19p and stabilizes U6 snRNA in catalytic
forms of the spliceosome containing U2, U5, and U6 snRNAs; null
mutant has splicing defect and arrests in G2/M; homologs
in human and C. elegans; K12867 pre-mRNA-splicing factor SYF1
Length=859
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 30/145 (20%), Positives = 63/145 (43%), Gaps = 19/145 (13%)
Query 3 LENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRH-------GLFKEALRLV---RLAI 52
+ A +++ ++++ Y I+DL ++ + L + ++AL + + +
Sbjct 463 ISTARELWTQSLKVPYPYIEDLEEIYLNWADRELDKEGVERAFSILEDALHVPTNPEILL 522
Query 53 SSSRKGQDTKGAQGRLFRSVKLWSLAADVEEMFGS---------IETVRACYDKMFQLKV 103
+ G AQ LF S+++WS D E + + Y+ + L++
Sbjct 523 EKYKNGHRKIPAQTVLFNSLRIWSKYIDYLEAYCPKDANSSDKIFNKTKMAYNTVIDLRL 582
Query 104 ITPQLVINYAHYLEERRYYEESFRV 128
ITP + N+A +L+ ESF+V
Sbjct 583 ITPAMAENFALFLQNHYEVMESFQV 607
> cel:M03F8.3 hypothetical protein; K12869 crooked neck
Length=747
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 33/141 (23%), Positives = 59/141 (41%), Gaps = 24/141 (17%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLR-------HGLFKEALRLVRLAIS 53
G+++ A VFERA+ +R+I S+W + EM +R +F A+ ++ A+
Sbjct 100 GEIQRARSVFERALDVDHRSI----SIWLQYAEMEMRCKQINHARNVFDRAITIMPRAMQ 155
Query 54 SSRKG-------QDTKGAQGRLFRSV------KLWSLAADVEEMFGSIETVRACYDKMFQ 100
K ++ GA+ R + + W + E + I+ R+ Y +
Sbjct 156 FWLKYSYMEEVIENIPGARQIFERWIEWEPPEQAWQTYINFELRYKEIDRARSVYQRFLH 215
Query 101 LKVITPQLVINYAHYLEERRY 121
+ I Q I YA + E Y
Sbjct 216 VHGINVQNWIKYAKFEERNGY 236
> hsa:51340 CRNKL1, CLF, CRN, Clf1, HCRN, MSTP021, SYF3; crooked
neck pre-mRNA splicing factor-like 1 (Drosophila); K12869
crooked neck
Length=848
Score = 32.3 bits (72), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 21/99 (21%), Positives = 44/99 (44%), Gaps = 17/99 (17%)
Query 2 DLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQDT 61
+++ A ++ERA+ YR I ++W + EM +++ A + AI++
Sbjct 257 EIQRARSIYERALDVDYRNI----TLWLKYAEMEMKNRQVNHARNIWDRAITT------- 305
Query 62 KGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQ 100
L R + W +EEM G++ R +++ +
Sbjct 306 ------LPRVNQFWYKYTYMEEMLGNVAGARQVFERWME 338
> mmu:66877 Crnkl1, 1200013P10Rik, 5730590A01Rik, C80326, crn;
Crn, crooked neck-like 1 (Drosophila); K12869 crooked neck
Length=690
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 21/99 (21%), Positives = 44/99 (44%), Gaps = 17/99 (17%)
Query 2 DLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQDT 61
+++ A ++ERA+ YR I ++W + EM +++ A + AI++
Sbjct 96 EIQRARSIYERALDVDYRNI----TLWLKYAEMEMKNRQVNHARNIWDRAITT------- 144
Query 62 KGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQ 100
L R + W +EEM G++ R +++ +
Sbjct 145 ------LPRVNQFWYKYTYMEEMLGNVAGARQVFERWME 177
> sce:YLR117C CLF1, NTC77, SYF3; Clf1p; K12869 crooked neck
Length=687
Score = 31.2 bits (69), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 27/96 (28%), Positives = 39/96 (40%), Gaps = 17/96 (17%)
Query 2 DLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQDT 61
D+ A +FERA+ I +W ++ L+ A L+ AIS+
Sbjct 80 DMRRARSIFERALLVDSSFI----PLWIRYIDAELKVKCINHARNLMNRAIST------- 128
Query 62 KGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDK 97
L R KLW VEE ++E VR+ Y K
Sbjct 129 ------LPRVDKLWYKYLIVEESLNNVEIVRSLYTK 158
> xla:779090 pdcd11, alg-4, alg4, nfbp, rrp5; programmed cell
death 11; K14792 rRNA biogenesis protein RRP5
Length=1812
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQD 60
GD E A +FE + ++Y DL SV+ ++M ++HG KE + I S +
Sbjct 1716 GDTERAKALFESTL-SSYPKRTDLWSVY---IDMMVKHGSQKEVRDIFERVIHLSLAAKK 1771
Query 61 TKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQ 100
K R + E+ GS E+V+A +K Q
Sbjct 1772 IKFFFKRYL----------EYEKKHGSTESVQAVKEKALQ 1801
> mmu:328110 Prpf39, FLJ11128, MGC37077, Srcs1; PRP39 pre-mRNA
processing factor 39 homolog (yeast); K13217 pre-mRNA-processing
factor 39
Length=665
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 29/118 (24%), Positives = 54/118 (45%), Gaps = 18/118 (15%)
Query 2 DLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKG--Q 59
+++ + +V+ R +QA ++D +W + KE L +++ +G +
Sbjct 142 NIKQSDEVYRRGLQAIPLSVD----LWIHYIN------FLKETLDPGDQETNTTIRGTFE 191
Query 60 DTKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLE 117
A G FRS KLW + + E G++ V A YD++ I QL Y+H+ +
Sbjct 192 HAVLAAGTDFRSDKLWEMYINWENEQGNLREVTAVYDRILG---IPTQL---YSHHFQ 243
> mmu:225638 Alpk2, Gm549, Hak; alpha-kinase 2; K08868 alpha-kinase
Length=2144
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 53 SSSRKGQDTKGAQGRLFRSVKLWSLAADVEE 83
S+ ++ QD K A+G LF S W A + E
Sbjct 854 STDKRSQDGKSAEGLLFNSTFTWDTAKEASE 884
> dre:794079 pdcd11, MGC162501, cb680, im:7148359, sb:cb680, wu:fc68c10;
programmed cell death 11; K14792 rRNA biogenesis
protein RRP5
Length=1816
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 24/48 (50%), Gaps = 0/48 (0%)
Query 70 RSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLE 117
+ LWS+ D+ GS + VR +D++ L V ++ + YLE
Sbjct 1738 KRTDLWSVFIDLMVKHGSQKEVRELFDRVIHLSVSVKKIKFFFKRYLE 1785
> mmu:233064 Wdr62, 2310038K02Rik, MGC31423; WD repeat domain
62
Length=1524
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 34 MHLRHGLFKEALRLVRLAISSSRKGQDTKGAQGRL 68
+H F+EAL L R+ +SSS+ G + + AQ L
Sbjct 1421 LHRLQTAFQEALDLYRMLVSSSQLGPEQQQAQTEL 1455
> ath:AT3G08530 clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1703
Score = 28.9 bits (63), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 3/43 (6%)
Query 3 LENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEAL 45
L+N + ERAV+ +R +D SVW + + LR GL +A+
Sbjct 1100 LDNVRSI-ERAVEFAFRVEED--SVWSQVAKAQLREGLVSDAI 1139
> ath:AT5G17460 hypothetical protein
Length=309
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 39/84 (46%), Gaps = 11/84 (13%)
Query 8 KVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQDTKGAQGR 67
K+FE + YRTI +L E E+ + +LA+ ++ K + KG GR
Sbjct 170 KLFEFEEKPKYRTISELLKSENEPEELS-------PGKKARKLAVENALKKLN-KGPDGR 221
Query 68 LFRSVKLWSLAADVEEMFGSIETV 91
+W + +DV+ + G+ E +
Sbjct 222 Y---TNVWEVMSDVDILIGAFENI 242
> hsa:55015 PRPF39, FLJ11128, FLJ20666, FLJ45460, MGC149842, MGC149843;
PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae);
K13217 pre-mRNA-processing factor 39
Length=669
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 28/54 (51%), Gaps = 6/54 (11%)
Query 64 AQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYLE 117
A G FRS +LW + + E G++ V A YD++ I QL Y+H+ +
Sbjct 198 AAGTDFRSDRLWEMYINWENEQGNLREVTAIYDRILG---IPTQL---YSHHFQ 245
> tpv:TP01_0426 hypothetical protein
Length=480
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 15/66 (22%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query 57 KGQDTKGAQGRLFRSVKLWSLAADVEEMFGSIETVRACYDKMFQLKVITPQLVINYAHYL 116
K ++KG+ G+ F S++ +A++ +++TV CY F++ + + Y YL
Sbjct 118 KTTESKGSDGK-FMSLEKNRFSAEMRSFNTTLQTVYNCYFPYFKMIYTSLDRIRTYTEYL 176
Query 117 EERRYY 122
+ ++
Sbjct 177 TKSSFH 182
> xla:495937 ppil6; peptidylprolyl isomerase (cyclophilin)-like
6 (EC:5.2.1.8); K12739 peptidyl-prolyl cis-trans isomerase-like
6 [EC:5.2.1.8]
Length=305
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 89 ETVRACYDKMFQLKVITPQLVINYAHYLEERR 120
E ++ +D F V+TP L + HYL E++
Sbjct 38 EVLKRSFDAEFDHPVVTPLLDCEWQHYLSEKK 69
> xla:447694 ogt, MGC80426; O-linked N-acetylglucosamine (GlcNAc)
transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase); K09667 polypeptide N-acetylglucosaminyltransferase
[EC:2.4.1.-]
Length=1063
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 33/134 (24%), Positives = 57/134 (42%), Gaps = 17/134 (12%)
Query 1 GDLENAAKVFERAVQATYR---TIDDLASVWCEAVEMHLRHGLFKEALRLV-RLAISSSR 56
G++E A +++ +A++ +LASV + ++ +KEA+R+ A + S
Sbjct 341 GNIEEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSN 400
Query 57 KG------QDTKGAQGRLFRSVKLWSLAADVEEMF-------GSIETVRACYDKMFQLKV 103
G QD +GA R++++ AD G+I A Y +LK
Sbjct 401 MGNTLKEMQDVQGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIASYRTALKLKP 460
Query 104 ITPQLVINYAHYLE 117
P N AH L+
Sbjct 461 DFPDAYCNLAHCLQ 474
> mmu:108155 Ogt, 1110038P24Rik, 4831420N21Rik, AI115525, Ogtl;
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase);
K09667 polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1046
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 33/134 (24%), Positives = 57/134 (42%), Gaps = 17/134 (12%)
Query 1 GDLENAAKVFERAVQATYR---TIDDLASVWCEAVEMHLRHGLFKEALRLV-RLAISSSR 56
G++E A +++ +A++ +LASV + ++ +KEA+R+ A + S
Sbjct 341 GNIEEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSN 400
Query 57 KG------QDTKGAQGRLFRSVKLWSLAADVEEMF-------GSIETVRACYDKMFQLKV 103
G QD +GA R++++ AD G+I A Y +LK
Sbjct 401 MGNTLKEMQDVQGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIASYRTALKLKP 460
Query 104 ITPQLVINYAHYLE 117
P N AH L+
Sbjct 461 DFPDAYCNLAHCLQ 474
> hsa:8473 OGT, FLJ23071, HRNT1, MGC22921, O-GLCNAC; O-linked
N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase); K09667
polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1046
Score = 28.5 bits (62), Expect = 6.2, Method: Composition-based stats.
Identities = 33/134 (24%), Positives = 57/134 (42%), Gaps = 17/134 (12%)
Query 1 GDLENAAKVFERAVQATYR---TIDDLASVWCEAVEMHLRHGLFKEALRLV-RLAISSSR 56
G++E A +++ +A++ +LASV + ++ +KEA+R+ A + S
Sbjct 341 GNIEEAVRLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSN 400
Query 57 KG------QDTKGAQGRLFRSVKLWSLAADVEEMF-------GSIETVRACYDKMFQLKV 103
G QD +GA R++++ AD G+I A Y +LK
Sbjct 401 MGNTLKEMQDVQGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIASYRTALKLKP 460
Query 104 ITPQLVINYAHYLE 117
P N AH L+
Sbjct 461 DFPDAYCNLAHCLQ 474
> ath:AT5G55840 pentatricopeptide (PPR) repeat-containing protein
Length=1274
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 13 AVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRL 50
A+ TYR + SV+ + ++LR G+ +++L + RL
Sbjct 111 ALMTTYRLCNSNPSVYDILIRVYLREGMIQDSLEIFRL 148
> dre:337685 ogt.1, fm81g08, ogt, wu:fc12b01, wu:fm81g08; O-linked
N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl
transferase) 1;
K09667 polypeptide N-acetylglucosaminyltransferase [EC:2.4.1.-]
Length=1062
Score = 27.7 bits (60), Expect = 8.2, Method: Composition-based stats.
Identities = 33/134 (24%), Positives = 57/134 (42%), Gaps = 17/134 (12%)
Query 1 GDLENAAKVFERAVQATYR---TIDDLASVWCEAVEMHLRHGLFKEALRLV-RLAISSSR 56
G++E A +++ +A++ +LASV + ++ +KEA+R+ A + S
Sbjct 341 GNIEEAVQLYRKALEVFPEFAAAHSNLASVLQQQGKLQEALMHYKEAIRISPTFADAYSN 400
Query 57 KG------QDTKGAQGRLFRSVKLWSLAADVEEMF-------GSIETVRACYDKMFQLKV 103
G QD +GA R++++ AD G+I A Y +LK
Sbjct 401 MGNTLKEMQDVQGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIASYRTALKLKP 460
Query 104 ITPQLVINYAHYLE 117
P N AH L+
Sbjct 461 DFPDAYCNLAHCLQ 474
> hsa:22984 PDCD11, ALG-4, ALG4, KIAA0185, NFBP, RRP5; programmed
cell death 11; K14792 rRNA biogenesis protein RRP5
Length=1871
Score = 27.7 bits (60), Expect = 8.3, Method: Composition-based stats.
Identities = 16/60 (26%), Positives = 29/60 (48%), Gaps = 8/60 (13%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEAL----RLVRLAISSSR 56
GD E A +FE + + D VW ++M ++HG K+ R++ L+++ R
Sbjct 1775 GDAERAKAIFENTLSTYPKRTD----VWSVYIDMTIKHGSQKDVRDIFERVIHLSLAPKR 1830
> ath:AT3G11130 clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1705
Score = 27.7 bits (60), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Query 3 LENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEAL 45
L + + ERAV+ +R +D +VW + + LR GL +A+
Sbjct 1099 LLDNVRSIERAVEFAFRVEED--AVWSQVAKAQLREGLVSDAI 1139
> ath:AT1G31790 pentatricopeptide (PPR) repeat-containing protein
Length=409
Score = 27.7 bits (60), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 32/61 (52%), Gaps = 6/61 (9%)
Query 1 GDLENAAKVFERAVQATYRTIDDLASVWCEAVEMHLRHGLFKEALRLVRLAISSSRKGQD 60
G +++A KVF+ + T S W V ++++G++ EA++L+ ++ K D
Sbjct 346 GKVKDAEKVFKSSKDETS------VSCWNAMVASYMQNGIYIEAIKLLYQMKATGIKAHD 399
Query 61 T 61
T
Sbjct 400 T 400
Lambda K H
0.324 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40