bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5118_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
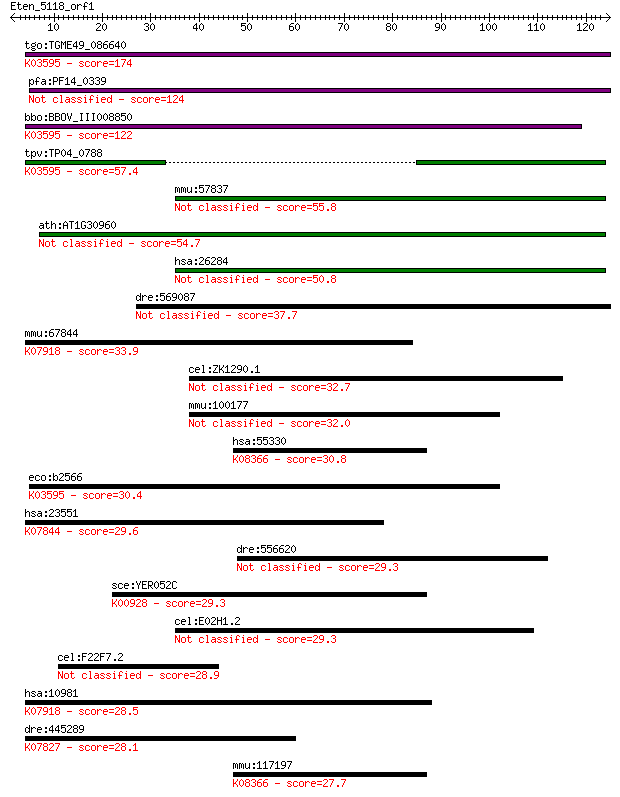
tgo:TGME49_086640 hypothetical protein ; K03595 GTP-binding pr... 174 5e-44
pfa:PF14_0339 conserved protein, unknown function 124 5e-29
bbo:BBOV_III008850 17.m07771; small GTP-binding protein domain... 122 3e-28
tpv:TP04_0788 hypothetical protein; K03595 GTP-binding protein... 57.4 1e-08
mmu:57837 Eral1, 2610524P08Rik, 9130407C09Rik, AU019798, Era, ... 55.8 3e-08
ath:AT1G30960 GTP-binding protein (ERG) 54.7 6e-08
hsa:26284 ERAL1, ERA, ERAL1A, H-ERA, HERA-A, HERA-B; Era G-pro... 50.8 1e-06
dre:569087 eral1, si:ch211-207c6.1; Era G-protein-like 1 (E. c... 37.7 0.010
mmu:67844 Rab32, 2810011A17Rik, AU022057; RAB32, member RAS on... 33.9 0.15
cel:ZK1290.1 hypothetical protein 32.7 0.30
mmu:100177 Zmym6, 9330177P20Rik, AI593204, AI661340, D4Wsu24e,... 32.0 0.49
hsa:55330 CNO, BCAS4L, FLJ11230; cappuccino homolog (mouse); K... 30.8 1.1
eco:b2566 era, ECK2564, JW2550, sdgE; membrane-associated, 16S... 30.4 1.3
hsa:23551 RASD2, MGC:4834, Rhes, TEM2; RASD family, member 2; ... 29.6 2.5
dre:556620 fi40d09; wu:fi40d09 29.3 2.8
sce:YER052C HOM3, BOR1, SIL4; Aspartate kinase (L-aspartate 4-... 29.3 3.3
cel:E02H1.2 hypothetical protein 29.3 3.3
cel:F22F7.2 hypothetical protein 28.9 4.5
hsa:10981 RAB32; RAB32, member RAS oncogene family; K07918 Ras... 28.5 6.2
dre:445289 kras, wu:fa04e08, wu:fc14b12, wu:fc23g10, wu:fj89d1... 28.1 7.8
mmu:117197 Cno, 2610101N07Rik; cappuccino; K08366 cappuccino 27.7 8.5
> tgo:TGME49_086640 hypothetical protein ; K03595 GTP-binding
protein Era
Length=413
Score = 174 bits (442), Expect = 5e-44, Method: Compositional matrix adjust.
Identities = 79/122 (64%), Positives = 98/122 (80%), Gaps = 1/122 (0%)
Query 4 IPLILCLNKIDLASHPKWIHAREKEFLSHGQFQNTFHISAKYKRGTDSLLDYLVSEAIPR 63
IP+ LC+NK+DLASHPKW++AR KEF +HG F+ FH+SAK RG LLD+LVS A
Sbjct 241 IPVFLCINKVDLASHPKWVYARAKEFQAHGHFEKIFHLSAKNSRGLQPLLDHLVSRAKRS 300
Query 64 PWAFPSEMRTSLSKVEQVEEFIRTYLFCWFNKDVPYRIDQQTVGWTYR-DGYVVIEHELV 122
WA+P EM+T+L KV+QV E I+TYLFCWFNKDVPYRI+QQT+GWT R DG ++IE EL+
Sbjct 301 VWAYPPEMKTTLPKVQQVAELIKTYLFCWFNKDVPYRIEQQTIGWTPRFDGSLIIEQELL 360
Query 123 VK 124
VK
Sbjct 361 VK 362
> pfa:PF14_0339 conserved protein, unknown function
Length=504
Score = 124 bits (312), Expect = 5e-29, Method: Composition-based stats.
Identities = 55/122 (45%), Positives = 86/122 (70%), Gaps = 2/122 (1%)
Query 5 PLILCLNKIDLASHPKWIHAREKEFLSHGQFQNTFHISAKYKRGTDSLLDYLVS-EAIPR 63
P+IL LNK+DL +H KW +AR KE++++G F N F ISAKY +G + LL+Y++ + +
Sbjct 337 PVILVLNKVDLCTHNKWANARAKEYMNNGIFDNIFFISAKYNKGIEELLNYIIKYKTKNQ 396
Query 64 PWAFPSEMRTSLSKVEQVEEFIRTYLFCWFNKDVPYRIDQQTVGWTYR-DGYVVIEHELV 122
W +P + T+LSKV+ VE+ I TY++CWFNKDVPY++ + W D ++IE++++
Sbjct 397 YWVYPKDAITTLSKVQIVEQLINTYIYCWFNKDVPYKVKHHMLSWCVNLDNSLIIEYQII 456
Query 123 VK 124
VK
Sbjct 457 VK 458
> bbo:BBOV_III008850 17.m07771; small GTP-binding protein domain
containing protein; K03595 GTP-binding protein Era
Length=340
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 55/116 (47%), Positives = 81/116 (69%), Gaps = 1/116 (0%)
Query 4 IPLILCLNKIDLASHPKWIHAREKEFLSHGQFQNTFHISAKYKRGTDSLLDYLVSEAIPR 63
IP+ L LNKIDL H KW+ R +E +HG F + F+ SAK+ G + ++ +L S A
Sbjct 161 IPVALVLNKIDLVKHKKWVKCRTRELKNHGAFADIFYTSAKHGIGGEHIITFLKSMAKSG 220
Query 64 PWAFPSEMRTSLSKVEQVEEFIRTYLFCWFNKDVPYRIDQQTVGWTYRD-GYVVIE 118
WA+P +M T++S+V+ VE+ +RTYL+CWFNK++PY+I Q+ +GWT D G +VIE
Sbjct 221 HWAYPPDMVTTMSRVQVVEQTVRTYLYCWFNKELPYQIGQKIIGWTRVDNGSLVIE 276
> tpv:TP04_0788 hypothetical protein; K03595 GTP-binding protein
Era
Length=276
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/40 (57%), Positives = 31/40 (77%), Gaps = 1/40 (2%)
Query 85 IRTYLFCWFNKDVPYRIDQQTVGWTY-RDGYVVIEHELVV 123
++TY+FCWFNKDVPY+IDQ +GWTY +G + IE + V
Sbjct 197 LKTYIFCWFNKDVPYKIDQSIIGWTYAENGTLNIEMSVKV 236
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 4 IPLILCLNKIDLASHPKWIHAREKEFLSH 32
+P+ L LNKIDL SH KW+ +R KE ++
Sbjct 172 VPIALVLNKIDLVSHKKWVKSRVKELKTY 200
> mmu:57837 Eral1, 2610524P08Rik, 9130407C09Rik, AU019798, Era,
M-ERA, MERA-S, MERA-W; Era (G-protein)-like 1 (E. coli)
Length=437
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 49/90 (54%), Gaps = 1/90 (1%)
Query 35 FQNTFHISAKYKRGTDSLLDYLVSEAIPRPWAFPSEMRTSLSKVEQVEEFIRTYLFCWFN 94
FQ F +SA + ++L YL+++A P PW F S + TS + E IR L +
Sbjct 302 FQEIFMLSALNNKDVNTLKQYLLTQAQPGPWEFHSGVLTSQTPEEICANKIREKLLEYLP 361
Query 95 KDVPYRIDQQTVGWTYR-DGYVVIEHELVV 123
++VPY + Q+TV W G +VI+ L+V
Sbjct 362 EEVPYGVQQKTVIWEEGPSGELVIQQNLLV 391
> ath:AT1G30960 GTP-binding protein (ERG)
Length=437
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 58/123 (47%), Gaps = 11/123 (8%)
Query 7 ILCLNKIDLASHPKWIHAREKEFLSHGQFQNTFHISAKYKRGTDSLLDYLVSEAIPRPW- 65
+LC+NK+DL K + +EF ++ F IS G L YL+ +A+ +PW
Sbjct 275 VLCMNKVDLVEKKKDLLKVAEEFQDLPAYERYFMISGLKGSGVKDLSQYLMDQAVKKPWE 334
Query 66 ----AFPSEMRTSLSKVEQVEEFIRTYLFCWFNKDVPYRIDQQTVGWT-YRDGYVVIEHE 120
E+ ++S E +R L ++++PY ++ + V W RDG + IE
Sbjct 335 EDAFTMSEEVLKNISL-----EVVRERLLDHVHQEIPYGLEHRLVDWKELRDGSLRIEQH 389
Query 121 LVV 123
L+
Sbjct 390 LIT 392
> hsa:26284 ERAL1, ERA, ERAL1A, H-ERA, HERA-A, HERA-B; Era G-protein-like
1 (E. coli)
Length=437
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 47/90 (52%), Gaps = 1/90 (1%)
Query 35 FQNTFHISAKYKRGTDSLLDYLVSEAIPRPWAFPSEMRTSLSKVEQVEEFIRTYLFCWFN 94
F+ F +SA + +L YL+++A P PW + S + TS + E IR L
Sbjct 302 FKEIFMLSALSQEDVKTLKQYLLTQAQPGPWEYHSAVLTSQTPEEICANIIREKLLEHLP 361
Query 95 KDVPYRIDQQTVGWTYR-DGYVVIEHELVV 123
++VPY + Q+T W G +VI+ +L+V
Sbjct 362 QEVPYNVQQKTAVWEEGPGGELVIQQKLLV 391
> dre:569087 eral1, si:ch211-207c6.1; Era G-protein-like 1 (E.
coli)
Length=447
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 31/102 (30%), Positives = 46/102 (45%), Gaps = 4/102 (3%)
Query 27 KEFLSHG---QFQNTFHISAKYKRGTDSLLDYLVSEAIPRPWAFPSEMRTSLSKVEQVEE 83
K SHG F++ F +S+ ++L YL A P W + SE+ T S +
Sbjct 301 KALKSHGGWPHFKDVFMLSSIDHEDVETLKRYLFVAAKPCQWQYHSEVLTDQSPEDVCFN 360
Query 84 FIRTYLFCWFNKDVPYRIDQQTVGWT-YRDGYVVIEHELVVK 124
IR L K+VPY + Q+ W DG + I +L V+
Sbjct 361 TIREKLLQNLPKEVPYTMTQEIEVWKESEDGVLDISIKLYVQ 402
> mmu:67844 Rab32, 2810011A17Rik, AU022057; RAB32, member RAS
oncogene family; K07918 Ras-related protein Rab-32
Length=223
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 33/80 (41%), Gaps = 1/80 (1%)
Query 4 IPLILCLNKIDLASHPKWIHAREKEFLSHGQFQNTFHISAKYKRGTDSLLDYLVSEAIPR 63
IP +L NK D ++ +F F F SAK D +LV +
Sbjct 134 IPAVLLANKCDQKKDNSQSPSQMDQFCKDHGFTGWFETSAKDNINIDEATRFLVENMLAN 193
Query 64 PWAFPSEMRTSLSKVEQVEE 83
+FPSE L +++ VEE
Sbjct 194 QQSFPSE-EIDLDRIKLVEE 212
> cel:ZK1290.1 hypothetical protein
Length=208
Score = 32.7 bits (73), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 35/80 (43%), Gaps = 9/80 (11%)
Query 38 TFHISAKYKRGT-DSLLDYLVSEAIP--RPWAFPSEMRTSLSKVEQVEEFIRTYLFCWFN 94
TF S + G DS + + +AI +PW +M+ LS E +E WFN
Sbjct 129 TFSRSNLHGEGVPDSFREATLPDAIKQKKPWTLMFKMKDGLSAFETGDET------GWFN 182
Query 95 KDVPYRIDQQTVGWTYRDGY 114
D+P+ T + R G+
Sbjct 183 NDIPWNGTATTQSFNIRGGW 202
> mmu:100177 Zmym6, 9330177P20Rik, AI593204, AI661340, D4Wsu24e,
Zfp258; zinc finger, MYM-type 6
Length=1249
Score = 32.0 bits (71), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 32/68 (47%), Gaps = 12/68 (17%)
Query 38 TFHISAKY---KRGTDSLLDYLVSEAIPRP-WAFPSEMRTSLSKVEQVEEFIRTYLFCWF 93
T ISAK ++GTD+L P+P +A + R S +E + RT LFC
Sbjct 170 TSGISAKCSMCQKGTDNL--------APKPLYALGNSSRLSAEMIETTNDSGRTELFCSI 221
Query 94 NKDVPYRI 101
N YRI
Sbjct 222 NCLSAYRI 229
> hsa:55330 CNO, BCAS4L, FLJ11230; cappuccino homolog (mouse);
K08366 cappuccino
Length=217
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 3/40 (7%)
Query 47 RGTDSLLDYLVSEAIPRPWAFPSEMRTSLSKVEQVEEFIR 86
RG S ++VSE +PR A +EMR S+++++E F+R
Sbjct 107 RGDSS---HVVSEGVPRIHAKAAEMRRIYSRIDRLEAFVR 143
> eco:b2566 era, ECK2564, JW2550, sdgE; membrane-associated, 16S
rRNA-binding GTPase; K03595 GTP-binding protein Era
Length=301
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 42/98 (42%), Gaps = 2/98 (2%)
Query 5 PLILCLNKIDLASHPKWIHAREKEFLSHGQFQNTFHISAKYKRGTDSLLDYLVSEAIPRP 64
P+IL +NK+D + + S F + ISA+ D++ +V + +P
Sbjct 118 PVILAVNKVDNVQEKADLLPHLQFLASQMNFLDIVPISAETGLNVDTIA-AIVRKHLPEA 176
Query 65 WA-FPSEMRTSLSKVEQVEEFIRTYLFCWFNKDVPYRI 101
FP + T S+ E IR L + ++PY +
Sbjct 177 THHFPEDYITDRSQRFMASEIIREKLMRFLGAELPYSV 214
> hsa:23551 RASD2, MGC:4834, Rhes, TEM2; RASD family, member 2;
K07844 RASD family, member 2
Length=266
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 31/75 (41%), Gaps = 6/75 (8%)
Query 4 IPLILCLNKIDLASHPKWIHAREKEFLSHGQFQNT-FHISAKYKRGTDSLLDYLVSEAIP 62
+P+++C NK D + + E E L G F +SAK D + L S A
Sbjct 133 LPMVICGNKNDHGELCRQVPTTEAELLVSGDENCAYFEVSAKKNTNVDEMFYVLFSMA-- 190
Query 63 RPWAFPSEMRTSLSK 77
P EM +L +
Sbjct 191 ---KLPHEMSPALHR 202
> dre:556620 fi40d09; wu:fi40d09
Length=2242
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 30/64 (46%), Gaps = 6/64 (9%)
Query 48 GTDSLLDYLVSEAIPRPWAFPSEMRTSLSKVEQVEEFIRTYLFCWFNKDVPYRIDQQTVG 107
GT L+Y +SE P P SE + +KV + ++++ + ++DQQT
Sbjct 1338 GTSGSLEYSISEEDPGPLRSDSEGKIGGTKVSRTFSYLKSKM------SKKNKVDQQTHN 1391
Query 108 WTYR 111
YR
Sbjct 1392 LVYR 1395
> sce:YER052C HOM3, BOR1, SIL4; Aspartate kinase (L-aspartate
4-P-transferase); cytoplasmic enzyme that catalyzes the first
step in the common pathway for methionine and threonine biosynthesis;
expression regulated by Gcn4p and the general control
of amino acid synthesis (EC:2.7.2.4); K00928 aspartate
kinase [EC:2.7.2.4]
Length=527
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 26/67 (38%), Positives = 32/67 (47%), Gaps = 9/67 (13%)
Query 22 IHAREKEFLSHGQFQNTFHISAKYKRGTD--SLLDYLVSEAIPRPWAFPSEMRTSLSKVE 79
IH+ +K LSHG F I KYK D S + VS A+P P A SL +
Sbjct 367 IHSNKKT-LSHGFLAQIFTILDKYKLVVDLISTSEVHVSMALPIPDA------DSLKSLR 419
Query 80 QVEEFIR 86
Q EE +R
Sbjct 420 QAEEKLR 426
> cel:E02H1.2 hypothetical protein
Length=394
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/74 (25%), Positives = 30/74 (40%), Gaps = 0/74 (0%)
Query 35 FQNTFHISAKYKRGTDSLLDYLVSEAIPRPWAFPSEMRTSLSKVEQVEEFIRTYLFCWFN 94
F+ F +S+ G D L D+L+S + W M T S + + IR +
Sbjct 253 FERVFFVSSLNGEGIDELRDHLMSISPQGEWKMQDGMPTGESAQQLCIDSIRAAVLDTTP 312
Query 95 KDVPYRIDQQTVGW 108
DV Y + + W
Sbjct 313 SDVAYTVQIRISEW 326
> cel:F22F7.2 hypothetical protein
Length=422
Score = 28.9 bits (63), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 11 NKIDLASHPKWIHAREKEFLSHGQFQNTFHISA 43
N+ID+A P+WI E ++ + N + +SA
Sbjct 111 NQIDVAGEPEWIERMEAKYGQMAKNNNVYIVSA 143
> hsa:10981 RAB32; RAB32, member RAS oncogene family; K07918 Ras-related
protein Rab-32
Length=225
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 35/84 (41%), Gaps = 1/84 (1%)
Query 4 IPLILCLNKIDLASHPKWIHAREKEFLSHGQFQNTFHISAKYKRGTDSLLDYLVSEAIPR 63
IP +L NK D ++ +F F F SAK + +LV + +
Sbjct 136 IPAVLLANKCDQNKDSSQSPSQVDQFCKEHGFAGWFETSAKDNINIEEAARFLVEKILVN 195
Query 64 PWAFPSEMRTSLSKVEQVEEFIRT 87
+FP+E + K++ +E +R
Sbjct 196 HQSFPNE-ENDVDKIKLDQETLRA 218
> dre:445289 kras, wu:fa04e08, wu:fc14b12, wu:fc23g10, wu:fj89d12,
zgc:85725; v-Ki-ras2 Kirsten rat sarcoma viral oncogene
homolog; K07827 GTPase KRas
Length=188
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 28/56 (50%), Gaps = 2/56 (3%)
Query 4 IPLILCLNKIDLASHPKWIHAREKEFLSHGQFQNTFHISAKYKRGTDSLLDYLVSE 59
+P++L NK DL SH + +++ + L+ SAK ++G D LV E
Sbjct 109 VPMVLVGNKCDLQSHN--VDSKQAQDLARSYGIPFIETSAKTRQGVDDAFYTLVRE 162
> mmu:117197 Cno, 2610101N07Rik; cappuccino; K08366 cappuccino
Length=215
Score = 27.7 bits (60), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Query 47 RGTDSLLDYLVSEAIPRPWAFPSEMRTSLSKVEQVEEFIR 86
RG S ++V E +PR A +EMR +++++E F+R
Sbjct 106 RGDSS---HVVGEGVPRIHAKAAEMRRIYGRIDKLEAFVR 142
Lambda K H
0.324 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069971060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40