bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5112_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
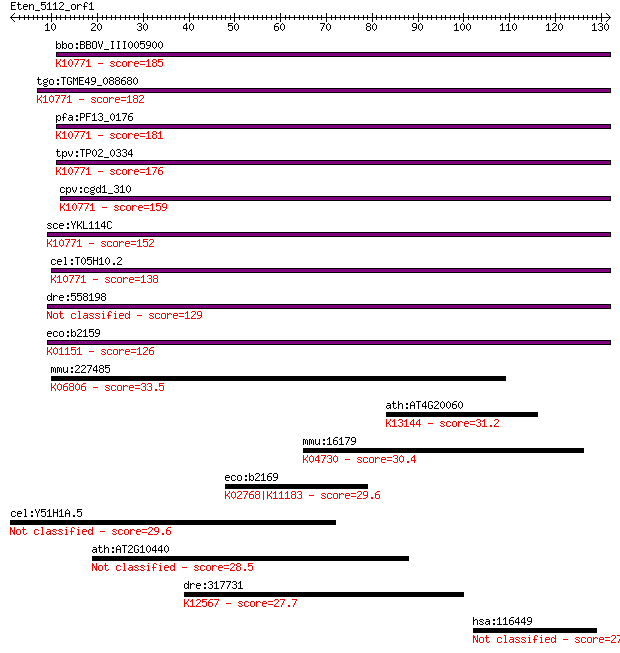
bbo:BBOV_III005900 17.m07523; apurinic endonuclease (APN1) fam... 185 3e-47
tgo:TGME49_088680 endonuclease V, putative (EC:3.1.21.2); K107... 182 2e-46
pfa:PF13_0176 apn1; apurinic/apyrimidinic endonuclease Apn1 (E... 181 4e-46
tpv:TP02_0334 apurinic/apyrimidinic endonuclease; K10771 AP en... 176 1e-44
cpv:cgd1_310 AP endonuclease of the TIM barrel fold, possible ... 159 2e-39
sce:YKL114C APN1; Apn1p (EC:4.2.99.18); K10771 AP endonuclease... 152 4e-37
cel:T05H10.2 apn-1; APurinic/apyrimidinic endoNuclease family ... 138 5e-33
dre:558198 APurinic/apyrimidinic endoNuclease family member (a... 129 2e-30
eco:b2159 nfo, ECK2152, JW2146; endonuclease IV with intrinsic... 126 2e-29
mmu:227485 Cdh19; cadherin 19, type 2; K06806 cadherin 19, type 2 33.5
ath:AT4G20060 EMB1895 (EMBRYO DEFECTIVE 1895); binding; K13144... 31.2 0.98
mmu:16179 Irak1, AA408924, IRAK, IRAK-1, IRAK1-S, Il1rak, Plpk... 30.4 1.5
eco:b2169 fruB, ECK2162, fpr, fruF, JW2156; fused fructose-spe... 29.6 2.4
cel:Y51H1A.5 hda-6; Histone DeAcetylase family member (hda-6) 29.6 2.6
ath:AT2G10440 hypothetical protein 28.5 4.9
dre:317731 ttna, fi20g08, si:busm1-258d18.1, si:dz258d18.1, tt... 27.7 8.7
hsa:116449 CLNK, MGC35197, MIST; cytokine-dependent hematopoie... 27.7 8.8
> bbo:BBOV_III005900 17.m07523; apurinic endonuclease (APN1) family
protein; K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=374
Score = 185 bits (470), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 87/121 (71%), Positives = 97/121 (80%), Gaps = 0/121 (0%)
Query 11 VLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIRN 70
VLPHGSYLIN+ANPD K+ +Y+ +DD+QRCE LGI LYN HPGSTVG C K EGIRN
Sbjct 148 VLPHGSYLINIANPDYEKRMKSYHHFVDDIQRCEKLGITLYNFHPGSTVGMCEKPEGIRN 207
Query 71 IAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHLFA 130
IA +N A K+T +VLEN AGQKNV+GS FEDLRDII LVENKDRV VCLDTCHLFA
Sbjct 208 IANCINMAMKETSSAKIVLENAAGQKNVIGSTFEDLRDIINLVENKDRVAVCLDTCHLFA 267
Query 131 A 131
A
Sbjct 268 A 268
> tgo:TGME49_088680 endonuclease V, putative (EC:3.1.21.2); K10771
AP endonuclease 1 [EC:4.2.99.18]
Length=667
Score = 182 bits (463), Expect = 2e-46, Method: Composition-based stats.
Identities = 83/125 (66%), Positives = 103/125 (82%), Gaps = 0/125 (0%)
Query 7 GQVQVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEE 66
G +LPHGSYLIN+ANPD AK++ +YNA LDDLQRCE +G+ Y HPGSTVG+C+KEE
Sbjct 442 GPEHILPHGSYLINLANPDAAKRKVSYNAFLDDLQRCEQIGVHRYVFHPGSTVGQCTKEE 501
Query 67 GIRNIAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTC 126
IR+IA +N+A TK V ++LENMAGQKNV+ S+FEDLRDII LV+ KDR+GVCLDTC
Sbjct 502 SIRHIAECLNKAIAATKSVTILLENMAGQKNVLCSEFEDLRDIIALVDRKDRIGVCLDTC 561
Query 127 HLFAA 131
HL++A
Sbjct 562 HLYSA 566
> pfa:PF13_0176 apn1; apurinic/apyrimidinic endonuclease Apn1
(EC:4.2.99.18); K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=597
Score = 181 bits (460), Expect = 4e-46, Method: Composition-based stats.
Identities = 82/121 (67%), Positives = 102/121 (84%), Gaps = 0/121 (0%)
Query 11 VLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIRN 70
+LPHGSYLIN+ANPDK K++ +Y + LDD++RCE L IKLYN HPGSTVG+C+ +EGI+N
Sbjct 377 ILPHGSYLINLANPDKEKRDKSYLSFLDDIKRCEQLNIKLYNFHPGSTVGQCTVDEGIKN 436
Query 71 IAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHLFA 130
+A +N+ HK+T V++VLEN AGQKN VGSKFE LRDII LV +KDR+GVCLDTCH FA
Sbjct 437 VADCINKVHKETNNVIIVLENSAGQKNSVGSKFEHLRDIINLVHDKDRIGVCLDTCHTFA 496
Query 131 A 131
A
Sbjct 497 A 497
> tpv:TP02_0334 apurinic/apyrimidinic endonuclease; K10771 AP
endonuclease 1 [EC:4.2.99.18]
Length=422
Score = 176 bits (447), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 82/121 (67%), Positives = 98/121 (80%), Gaps = 0/121 (0%)
Query 11 VLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIRN 70
VLPH SYLINVANPD K++ A++ LDD+QRCE LGI LYN HPGST G C K EGI++
Sbjct 204 VLPHASYLINVANPDPEKRKRAFDNFLDDIQRCEVLGITLYNFHPGSTCGLCEKSEGIQH 263
Query 71 IAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHLFA 130
I+ +N+A + TK V +VLEN AGQKNV+GSKFEDLR II+ VE+K RVG+CLDTCHLFA
Sbjct 264 ISDCINKALEMTKGVTIVLENAAGQKNVIGSKFEDLRQIIDKVEDKSRVGICLDTCHLFA 323
Query 131 A 131
A
Sbjct 324 A 324
> cpv:cgd1_310 AP endonuclease of the TIM barrel fold, possible
bacterial horizontal transfer ; K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=364
Score = 159 bits (403), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 74/120 (61%), Positives = 95/120 (79%), Gaps = 1/120 (0%)
Query 12 LPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIRNI 71
+PHGSYLIN+ NPD+ K+ Y A D+L+RC+ALGIKLYN HPGSTVG+CSKEE I+ I
Sbjct 145 IPHGSYLINLGNPDEEKRSKMYGAFEDELKRCDALGIKLYNFHPGSTVGQCSKEESIQFI 204
Query 72 AAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHLFAA 131
+ +N+AH+ TK V+ VLEN A K VG +F++L +IIE V++K R+GVCLDTCHLFAA
Sbjct 205 SDCINKAHEQTKSVITVLENCAESK-CVGYRFKELSEIIERVKDKSRIGVCLDTCHLFAA 263
> sce:YKL114C APN1; Apn1p (EC:4.2.99.18); K10771 AP endonuclease
1 [EC:4.2.99.18]
Length=367
Score = 152 bits (383), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 67/123 (54%), Positives = 93/123 (75%), Gaps = 1/123 (0%)
Query 9 VQVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGI 68
VLPHG Y IN+ANPD+ K E +Y + +DDL RCE LGI LYN+HPGST+ + + +
Sbjct 78 TDVLPHGQYFINLANPDREKAEKSYESFMDDLNRCEQLGIGLYNLHPGSTL-KGDHQLQL 136
Query 69 RNIAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHL 128
+ +A+ +N+A K+TKFV +VLENMAG N+VGS DL+++I ++E+K R+GVC+DTCH
Sbjct 137 KQLASYLNKAIKETKFVKIVLENMAGTGNLVGSSLVDLKEVIGMIEDKSRIGVCIDTCHT 196
Query 129 FAA 131
FAA
Sbjct 197 FAA 199
> cel:T05H10.2 apn-1; APurinic/apyrimidinic endoNuclease family
member (apn-1); K10771 AP endonuclease 1 [EC:4.2.99.18]
Length=396
Score = 138 bits (347), Expect = 5e-33, Method: Composition-based stats.
Identities = 63/122 (51%), Positives = 88/122 (72%), Gaps = 0/122 (0%)
Query 10 QVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIR 69
Q++PHGSYL+N +P+ K E + A+LD+ QR E LGI +YN HPGSTVG+C KEE +
Sbjct 181 QIVPHGSYLMNAGSPEAEKLEKSRLAMLDECQRAEKLGITMYNFHPGSTVGKCEKEECMT 240
Query 70 NIAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHLF 129
IA ++ + T+ +++VLE MAGQ N +G FE+L+ II+ V+ K RVGVC+DTCH+F
Sbjct 241 TIAETIDFVVEKTENIILVLETMAGQGNSIGGTFEELKFIIDKVKVKSRVGVCIDTCHIF 300
Query 130 AA 131
A
Sbjct 301 AG 302
> dre:558198 APurinic/apyrimidinic endoNuclease family member
(apn-1)-like
Length=296
Score = 129 bits (324), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 59/123 (47%), Positives = 85/123 (69%), Gaps = 1/123 (0%)
Query 9 VQVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGI 68
V +LPHGSYL+N +P + + L+D+L RC+ALG+ +N HPG+ + + S E+ +
Sbjct 74 VHILPHGSYLMNCGSPKEDVFSKSQAMLVDELSRCDALGLSQFNFHPGAAM-DSSPEKCM 132
Query 69 RNIAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHL 128
IA A+N AH+ T V+ VLENM+GQ + +G +F +L+ II+ V +K RVGVCLDTCH
Sbjct 133 EKIAQAINHAHQQTPAVITVLENMSGQGSTIGGQFSELKGIIDRVRDKSRVGVCLDTCHA 192
Query 129 FAA 131
FAA
Sbjct 193 FAA 195
> eco:b2159 nfo, ECK2152, JW2146; endonuclease IV with intrinsic
3'-5' exonuclease activity (EC:3.1.21.2); K01151 deoxyribonuclease
IV [EC:3.1.21.2]
Length=285
Score = 126 bits (316), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 61/123 (49%), Positives = 84/123 (68%), Gaps = 0/123 (0%)
Query 9 VQVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGI 68
Q+LPH SYLIN+ +P E + +A +D++QRCE LG+ L N HPGS + + S+E+ +
Sbjct 64 AQILPHDSYLINLGHPVTEALEKSRDAFIDEMQRCEQLGLSLLNFHPGSHLMQISEEDCL 123
Query 69 RNIAAAVNRAHKDTKFVVVVLENMAGQKNVVGSKFEDLRDIIELVENKDRVGVCLDTCHL 128
IA ++N A T+ V V+EN AGQ + +G KFE L II+ VE+K RVGVC+DTCH
Sbjct 124 ARIAESINIALDKTQGVTAVIENTAGQGSNLGFKFEHLAAIIDGVEDKSRVGVCIDTCHA 183
Query 129 FAA 131
FAA
Sbjct 184 FAA 186
> mmu:227485 Cdh19; cadherin 19, type 2; K06806 cadherin 19, type
2
Length=770
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 24/105 (22%), Positives = 43/105 (40%), Gaps = 18/105 (17%)
Query 10 QVLPHGSYLINVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIR 69
++ P G+++I V D Y+A + LYN+ G G+
Sbjct 157 EMSPEGTFVIKVTANDADDPSTGYHARI------------LYNLERGQPYFSVEPTTGVI 204
Query 70 NIAAAVNRAHKDTKFVVVVLENMAGQ------KNVVGSKFEDLRD 108
I++ ++R +DT V++ ++M GQ V K D+ D
Sbjct 205 RISSKMDRELQDTYCVIIQAKDMLGQPGALSGTTTVSIKLSDIND 249
> ath:AT4G20060 EMB1895 (EMBRYO DEFECTIVE 1895); binding; K13144
integrator complex subunit 7
Length=1134
Score = 31.2 bits (69), Expect = 0.98, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 83 KFVVVVLENMAGQKNVVGSKFEDLRDIIELVEN 115
KF++V LEN+ G N++ +E ++ I E V +
Sbjct 510 KFLIVFLENLEGDDNLLSEIYEKVKHITEFVSS 542
> mmu:16179 Irak1, AA408924, IRAK, IRAK-1, IRAK1-S, Il1rak, Plpk,
mPLK; interleukin-1 receptor-associated kinase 1 (EC:2.7.11.1);
K04730 interleukin-1 receptor-associated kinase 1 [EC:2.7.11.1]
Length=750
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 37/64 (57%), Gaps = 8/64 (12%)
Query 65 EEGIRNIAAAVNRAHKDT-KFVVVVLENMAGQKNV--VGSKFEDLRDIIELVENKDRVGV 121
EE I+ AV+ DT F VV+LE +AGQ+ V G+K + L+D+IE + + GV
Sbjct 394 EEYIKTGRLAVD---TDTFSFGVVILETLAGQRAVRTQGAKTKYLKDLIE--DEAEEAGV 448
Query 122 CLDT 125
L +
Sbjct 449 TLKS 452
> eco:b2169 fruB, ECK2162, fpr, fruF, JW2156; fused fructose-specific
PTS enzymes: IIA component/HPr component (EC:2.7.1.69);
K02768 PTS system, fructose-specific IIA component [EC:2.7.1.69];
K11183 phosphocarrier protein FPr
Length=376
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 2/31 (6%)
Query 48 IKLYNIHPGSTVGECSKEEGIRNIAAAVNRA 78
+ + +IHPG G+ KEE IR +AAA+ +A
Sbjct 4 LSVQDIHPGEKAGD--KEEAIRQVAAALVQA 32
> cel:Y51H1A.5 hda-6; Histone DeAcetylase family member (hda-6)
Length=517
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 31/74 (41%), Gaps = 14/74 (18%)
Query 1 AFERPPGQVQVLPHGSYLI---NVANPDKAKQENAYNALLDDLQRCEALGIKLYNIHPGS 57
AF RPPG + G NVA KA +N L+ D Y++H G+
Sbjct 130 AFIRPPGHHAMPDEGCGFCIFNNVAIAAKAAIQNGQKVLIVD-----------YDVHAGN 178
Query 58 TVGECSKEEGIRNI 71
EC ++ G N+
Sbjct 179 GTQECVEQMGEGNV 192
> ath:AT2G10440 hypothetical protein
Length=845
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 36/72 (50%), Gaps = 3/72 (4%)
Query 19 INVANPDKAKQENAY---NALLDDLQRCEALGIKLYNIHPGSTVGECSKEEGIRNIAAAV 75
IN+ PD + Q ++Y ++L ++ + G+K+ NI PG + + KE R + V
Sbjct 653 INIVPPDMSSQIDSYEQLSSLESEVVSTTSSGLKVNNIAPGYALLQEIKETNGRLVETVV 712
Query 76 NRAHKDTKFVVV 87
+D+ +V
Sbjct 713 EICDEDSLGTIV 724
> dre:317731 ttna, fi20g08, si:busm1-258d18.1, si:dz258d18.1,
ttn, wu:fi04b11, wu:fi20g08; titin a; K12567 titin [EC:2.7.11.1]
Length=32757
Score = 27.7 bits (60), Expect = 8.7, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 29/63 (46%), Gaps = 6/63 (9%)
Query 39 DLQRCEALGIKLYNIHPGSTVGECSKEE--GIRNIAAAVNRAHKDTKFVVVVLENMAGQK 96
D+Q+C+AL + Y + VG CS + I+ + V R VV VL M K
Sbjct 6078 DVQKCDALDVGEYQCVVANEVGSCSSQSTLSIKEPPSFVERIEN----VVTVLGKMVEFK 6133
Query 97 NVV 99
VV
Sbjct 6134 CVV 6136
> hsa:116449 CLNK, MGC35197, MIST; cytokine-dependent hematopoietic
cell linker
Length=428
Score = 27.7 bits (60), Expect = 8.8, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 8/35 (22%)
Query 102 KFEDLRDIIE--------LVENKDRVGVCLDTCHL 128
KF+ + DIIE L++ KD+ GV CHL
Sbjct 381 KFDSVEDIIEHYKNFPIILIDGKDKTGVHRKQCHL 415
Lambda K H
0.318 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40