bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5098_orf1
Length=64
Score E
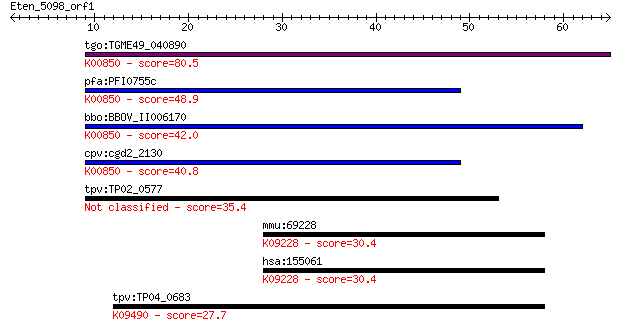
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_040890 phosphofructokinase, putative (EC:2.7.1.90);... 80.5 1e-15
pfa:PFI0755c 6-phosphofructokinase, putative; K00850 6-phospho... 48.9 4e-06
bbo:BBOV_II006170 18.m06510; 6-phosphofructokinase (EC:2.7.1.9... 42.0 4e-04
cpv:cgd2_2130 pyrophosphate-dependent phosphofructokinase (EC:... 40.8 0.001
tpv:TP02_0577 6-phosphofructokinase 35.4 0.046
mmu:69228 Zfp746, 2810407L07Rik, AI317225, Znf746; zinc finger... 30.4 1.3
hsa:155061 ZNF746, FLJ31413; zinc finger protein 746; K09228 K... 30.4 1.3
tpv:TP04_0683 heat shock protein 70; K09490 heat shock 70kDa p... 27.7 9.5
> tgo:TGME49_040890 phosphofructokinase, putative (EC:2.7.1.90);
K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=1399
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 38/56 (67%), Positives = 44/56 (78%), Gaps = 0/56 (0%)
Query 9 DALLMHLPNMKILLAELRRIIREAQSKGEMKKAQQQLMDVDSWAANEPDSWNWRLT 64
DALLMHLPNMKILLAELR +++EA +KGEMKKAQQ L DV + P+SW RLT
Sbjct 1026 DALLMHLPNMKILLAELRSVLKEADAKGEMKKAQQDLNDVTDTDSCPPESWGRRLT 1081
> pfa:PFI0755c 6-phosphofructokinase, putative; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=1418
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 21/40 (52%), Positives = 32/40 (80%), Gaps = 0/40 (0%)
Query 9 DALLMHLPNMKILLAELRRIIREAQSKGEMKKAQQQLMDV 48
DALLMHLP+MKILL+E+ I+ EA KG++ +A+ L+++
Sbjct 1088 DALLMHLPHMKILLSEISDILNEASEKGQLLEARNDLVNL 1127
> bbo:BBOV_II006170 18.m06510; 6-phosphofructokinase (EC:2.7.1.90);
K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=1337
Score = 42.0 bits (97), Expect = 4e-04, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 31/53 (58%), Gaps = 3/53 (5%)
Query 9 DALLMHLPNMKILLAELRRIIREAQSKGEMKKAQQQLMDVDSWAANEPDSWNW 61
D L++HLPN K +L ELR ++ EA + + ++A ++ D +EP S W
Sbjct 1008 DHLILHLPNTKSMLMELRSVLMEASAANKRREAVDSFLNYDK---SEPQSSEW 1057
> cpv:cgd2_2130 pyrophosphate-dependent phosphofructokinase (EC:2.7.1.11);
K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=1426
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 27/40 (67%), Gaps = 0/40 (0%)
Query 9 DALLMHLPNMKILLAELRRIIREAQSKGEMKKAQQQLMDV 48
DALL HLP+MKIL++E+ + R A+ EMK ++M +
Sbjct 1014 DALLSHLPDMKILISEINELRRFAEEHSEMKLFMSEMMSL 1053
> tpv:TP02_0577 6-phosphofructokinase
Length=1692
Score = 35.4 bits (80), Expect = 0.046, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 9 DALLMHLPNMKILLAELRRIIREAQSKGEMKKAQQQLMDVDSWA 52
D L++HLP+ K +L E+ + +A K + K A QQLM + ++
Sbjct 1319 DHLVLHLPSTKNMLQEIGTALTKANQKNQRKHAIQQLMSYNVYS 1362
> mmu:69228 Zfp746, 2810407L07Rik, AI317225, Znf746; zinc finger
protein 746; K09228 KRAB domain-containing zinc finger protein
Length=652
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 10/30 (33%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 28 IIREAQSKGEMKKAQQQLMDVDSWAANEPD 57
++ + + +GE++ +QQ + V++WAA +PD
Sbjct 184 LLMQIKQEGELQLQEQQALGVEAWAAGQPD 213
> hsa:155061 ZNF746, FLJ31413; zinc finger protein 746; K09228
KRAB domain-containing zinc finger protein
Length=645
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 10/30 (33%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 28 IIREAQSKGEMKKAQQQLMDVDSWAANEPD 57
++ + + +GE++ +QQ + V++WAA +PD
Sbjct 184 LLMQIKQEGELQLQEQQALGVEAWAAGQPD 213
> tpv:TP04_0683 heat shock protein 70; K09490 heat shock 70kDa
protein 5
Length=655
Score = 27.7 bits (60), Expect = 9.5, Method: Composition-based stats.
Identities = 13/51 (25%), Positives = 27/51 (52%), Gaps = 5/51 (9%)
Query 12 LMHLPNMKILLAELRRIIREAQ-----SKGEMKKAQQQLMDVDSWAANEPD 57
+M +++ + +RR ++E SK E+ K +++L D +W + PD
Sbjct 562 IMSKQSLENYIDSMRRTLKEDSVSSKLSKSEVSKLKEELDDASNWMGSHPD 612
Lambda K H
0.317 0.126 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2051595972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40