bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5094_orf1
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
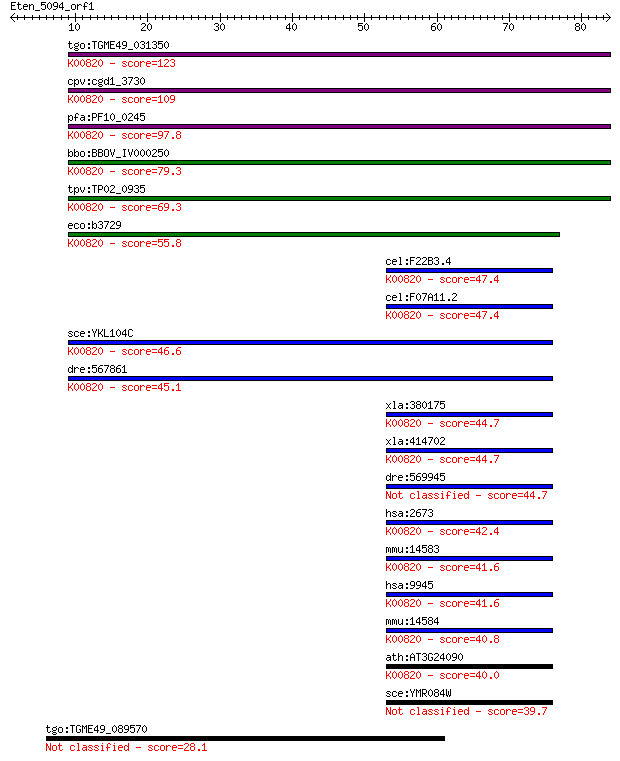
tgo:TGME49_031350 glucosamine--fructose-6-phosphate aminotrans... 123 1e-28
cpv:cgd1_3730 glucosamine-fructose-6-phosphate aminotransferas... 109 2e-24
pfa:PF10_0245 glucosamine-fructose-6-phosphate aminotransferas... 97.8 8e-21
bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate... 79.3 3e-15
tpv:TP02_0935 glucosamine-fructose-6-phosphate aminotransferas... 69.3 3e-12
eco:b3729 glmS, ECK3722, JW3707; L-glutamine:D-fructose-6-phos... 55.8 3e-08
cel:F22B3.4 hypothetical protein; K00820 glucosamine--fructose... 47.4 1e-05
cel:F07A11.2 hypothetical protein; K00820 glucosamine--fructos... 47.4 1e-05
sce:YKL104C GFA1; Gfa1p (EC:2.6.1.16); K00820 glucosamine--fru... 46.6 2e-05
dre:567861 gfpt1; glutamine-fructose-6-phosphate transaminase ... 45.1 5e-05
xla:380175 gfpt2, MGC53126, gfpt1; glutamine-fructose-6-phosph... 44.7 7e-05
xla:414702 gfpt1, MGC83201, gfat; glutamine--fructose-6-phosph... 44.7 7e-05
dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase ... 44.7 8e-05
hsa:2673 GFPT1, GFA, GFAT, GFAT_1, GFAT1, GFAT1m, GFPT; glutam... 42.4 4e-04
mmu:14583 Gfpt1, 2810423A18Rik, AI324119, AI449986, GFA, GFAT,... 41.6 6e-04
hsa:9945 GFPT2, FLJ10380, GFAT2; glutamine-fructose-6-phosphat... 41.6 6e-04
mmu:14584 Gfpt2, AI480523, GFAT2; glutamine fructose-6-phospha... 40.8 0.001
ath:AT3G24090 glutamine-fructose-6-phosphate transaminase (iso... 40.0 0.002
sce:YMR084W Putative protein of unknown function; YMR084W and ... 39.7 0.002
tgo:TGME49_089570 phosphatidylinositol transfer protein beta i... 28.1 7.3
> tgo:TGME49_031350 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing), putative (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=603
Score = 123 bits (309), Expect = 1e-28, Method: Composition-based stats.
Identities = 54/75 (72%), Positives = 61/75 (81%), Gaps = 0/75 (0%)
Query 9 YDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQHGGHHIGIAHTRWATHGGKT 68
YDSCG+T+I +GQL TTKFASRGTT D I++L+RE H G+HIGIAHTRWATHG KT
Sbjct 12 YDSCGITSISPEGQLVTTKFASRGTTCDSIEILRREGKHPHRGNHIGIAHTRWATHGSKT 71
Query 69 DTNAHPHMDWKKRIS 83
D NAHPH DWK RIS
Sbjct 72 DENAHPHHDWKDRIS 86
> cpv:cgd1_3730 glucosamine-fructose-6-phosphate aminotransferase
(EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=696
Score = 109 bits (272), Expect = 2e-24, Method: Composition-based stats.
Identities = 49/75 (65%), Positives = 59/75 (78%), Gaps = 1/75 (1%)
Query 9 YDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQHGGHHIGIAHTRWATHGGKT 68
YDSCGM+TI G+L TTK++S+ + D I+ LK ++ HG HHIGIAHTRWATHGGKT
Sbjct 111 YDSCGMSTIDDQGELITTKYSSK-ESGDSIERLKNDSELLHGNHHIGIAHTRWATHGGKT 169
Query 69 DTNAHPHMDWKKRIS 83
D NAHPH D+KKRIS
Sbjct 170 DFNAHPHQDYKKRIS 184
> pfa:PF10_0245 glucosamine-fructose-6-phosphate aminotransferase,
putative; K00820 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing) [EC:2.6.1.16]
Length=829
Score = 97.8 bits (242), Expect = 8e-21, Method: Composition-based stats.
Identities = 46/75 (61%), Positives = 53/75 (70%), Gaps = 1/75 (1%)
Query 9 YDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQHGGHHIGIAHTRWATHGGKT 68
YDSCGM+TI L TTK+AS TT D I+ LK + H HIGIAHTRWATHG KT
Sbjct 242 YDSCGMSTISNKNVLKTTKYASN-TTCDAIEKLKSNYLNSHKNDHIGIAHTRWATHGCKT 300
Query 69 DTNAHPHMDWKKRIS 83
D NAHPH+D+ +RIS
Sbjct 301 DENAHPHVDYGERIS 315
> bbo:BBOV_IV000250 21.m02906; glucosamine--fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=723
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 36/75 (48%), Positives = 46/75 (61%), Gaps = 0/75 (0%)
Query 9 YDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQHGGHHIGIAHTRWATHGGKT 68
YDSCG+ T+ DG + TK +S ++C L+ +H G IGIAHTRWAT G T
Sbjct 80 YDSCGIGTMRSDGSIEVTKCSSYKAPANCFDRLRERLVDRHLGSTIGIAHTRWATMGPPT 139
Query 69 DTNAHPHMDWKKRIS 83
D NAHPH D K R++
Sbjct 140 DENAHPHCDPKGRVA 154
> tpv:TP02_0935 glucosamine-fructose-6-phosphate aminotransferase;
K00820 glucosamine--fructose-6-phosphate aminotransferase
(isomerizing) [EC:2.6.1.16]
Length=810
Score = 69.3 bits (168), Expect = 3e-12, Method: Composition-based stats.
Identities = 33/75 (44%), Positives = 46/75 (61%), Gaps = 1/75 (1%)
Query 9 YDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQHGGHHIGIAHTRWATHGGKT 68
YDSCG+ T+ DGQL TK S T +D +LK + + H +GI HTRWAT G +
Sbjct 62 YDSCGVCTLH-DGQLKVTKCCSVETPADSFNILKDKVLATHPPSTVGIGHTRWATVGKLS 120
Query 69 DTNAHPHMDWKKRIS 83
+ N+HPH+D K ++
Sbjct 121 NKNSHPHVDLAKNLA 135
> eco:b3729 glmS, ECK3722, JW3707; L-glutamine:D-fructose-6-phosphate
aminotransferase (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=609
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 28/70 (40%), Positives = 40/70 (57%), Gaps = 10/70 (14%)
Query 9 YDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQ--HGGHHIGIAHTRWATHGG 66
YDS G+ + +G +T + + +Q+L + A HGG GIAHTRWATHG
Sbjct 29 YDSAGLAVVDAEGHMTRLRRLGK------VQMLAQAAEEHPLHGG--TGIAHTRWATHGE 80
Query 67 KTDTNAHPHM 76
++ NAHPH+
Sbjct 81 PSEVNAHPHV 90
> cel:F22B3.4 hypothetical protein; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=710
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 17/23 (73%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H GIAHTRWATHG D N+HPH
Sbjct 87 HCGIAHTRWATHGSPRDVNSHPH 109
> cel:F07A11.2 hypothetical protein; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=712
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 17/23 (73%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H GIAHTRWATHG D N+HPH
Sbjct 87 HCGIAHTRWATHGSPRDVNSHPH 109
> sce:YKL104C GFA1; Gfa1p (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=717
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/69 (37%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Query 9 YDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQHGG--HHIGIAHTRWATHGG 66
YDS G+ I GD +T + G S + + ++ P++ H GIAHTRWATHG
Sbjct 35 YDSTGIA-IDGDEADSTFIYKQIGKVSALKEEITKQNPNRDVTFVSHCGIAHTRWATHGR 93
Query 67 KTDTNAHPH 75
N HP
Sbjct 94 PEQVNCHPQ 102
> dre:567861 gfpt1; glutamine-fructose-6-phosphate transaminase
1 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 36/83 (43%), Gaps = 23/83 (27%)
Query 9 YDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQHGGH---------------- 52
YDS G+ GG+ + + S CI L+K+ Q
Sbjct 35 YDSAGVGIDGGNSK-------DWESNSKCIHLIKQTGKVQALDEEIQRQQDIDLDVEFDV 87
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H+GIAHTRWATHG + N+HP
Sbjct 88 HLGIAHTRWATHGVPSPVNSHPQ 110
> xla:380175 gfpt2, MGC53126, gfpt1; glutamine-fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 16/23 (69%), Positives = 19/23 (82%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H+GIAHTRWATHG + TN+HP
Sbjct 88 HLGIAHTRWATHGEPSPTNSHPQ 110
> xla:414702 gfpt1, MGC83201, gfat; glutamine--fructose-6-phosphate
transaminase 1; K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 44.7 bits (104), Expect = 7e-05, Method: Composition-based stats.
Identities = 16/23 (69%), Positives = 19/23 (82%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H+GIAHTRWATHG + TN+HP
Sbjct 88 HLGIAHTRWATHGEPSPTNSHPQ 110
> dre:569945 gfpt2; glutamine-fructose-6-phosphate transaminase
2 (EC:2.6.1.16)
Length=681
Score = 44.7 bits (104), Expect = 8e-05, Method: Composition-based stats.
Identities = 16/23 (69%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H GIAHTRWATHG + N+HPH
Sbjct 84 HFGIAHTRWATHGEPSPLNSHPH 106
> hsa:2673 GFPT1, GFA, GFAT, GFAT_1, GFAT1, GFAT1m, GFPT; glutamine--fructose-6-phosphate
transaminase 1 (EC:2.6.1.16); K00820
glucosamine--fructose-6-phosphate aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=681
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 15/23 (65%), Positives = 18/23 (78%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H+GIAHTRWATHG + N+HP
Sbjct 88 HLGIAHTRWATHGEPSPVNSHPQ 110
> mmu:14583 Gfpt1, 2810423A18Rik, AI324119, AI449986, GFA, GFAT,
GFAT1, GFAT1m, Gfpt; glutamine fructose-6-phosphate transaminase
1 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=681
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 15/23 (65%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H+GIAHTRWATHG N+HP
Sbjct 88 HLGIAHTRWATHGEPNPVNSHPQ 110
> hsa:9945 GFPT2, FLJ10380, GFAT2; glutamine-fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 15/23 (65%), Positives = 17/23 (73%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H GIAHTRWATHG + N+HP
Sbjct 85 HFGIAHTRWATHGVPSAVNSHPQ 107
> mmu:14584 Gfpt2, AI480523, GFAT2; glutamine fructose-6-phosphate
transaminase 2 (EC:2.6.1.16); K00820 glucosamine--fructose-6-phosphate
aminotransferase (isomerizing) [EC:2.6.1.16]
Length=682
Score = 40.8 bits (94), Expect = 0.001, Method: Composition-based stats.
Identities = 15/23 (65%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H GIAHTRWATHG N+HP
Sbjct 85 HFGIAHTRWATHGVPNAVNSHPQ 107
> ath:AT3G24090 glutamine-fructose-6-phosphate transaminase (isomerizing)/
sugar binding / transaminase (EC:2.6.1.16); K00820
glucosamine--fructose-6-phosphate aminotransferase (isomerizing)
[EC:2.6.1.16]
Length=695
Score = 40.0 bits (92), Expect = 0.002, Method: Composition-based stats.
Identities = 15/23 (65%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H GIAHTRWATHG N+HP
Sbjct 85 HAGIAHTRWATHGEPAPRNSHPQ 107
> sce:YMR084W Putative protein of unknown function; YMR084W and
adjacent ORF YMR085W are merged in related strains
Length=262
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 16/23 (69%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 53 HIGIAHTRWATHGGKTDTNAHPH 75
H GIAHTR ATHGG N HPH
Sbjct 80 HCGIAHTRRATHGGLRRANCHPH 102
> tgo:TGME49_089570 phosphatidylinositol transfer protein beta
isoform, putative
Length=1245
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 14/55 (25%), Positives = 23/55 (41%), Gaps = 0/55 (0%)
Query 6 PGRYDSCGMTTIGGDGQLTTTKFASRGTTSDCIQLLKREAPSQHGGHHIGIAHTR 60
P +Y C + + TK + G +DC+++LK E + G H R
Sbjct 14 PEQYKRCQLYMVAKASLEDATKAEAEGAANDCMEILKNEEYTNEEGDTGQYTHKR 68
Lambda K H
0.316 0.132 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044675024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40