bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_5045_orf1
Length=86
Score E
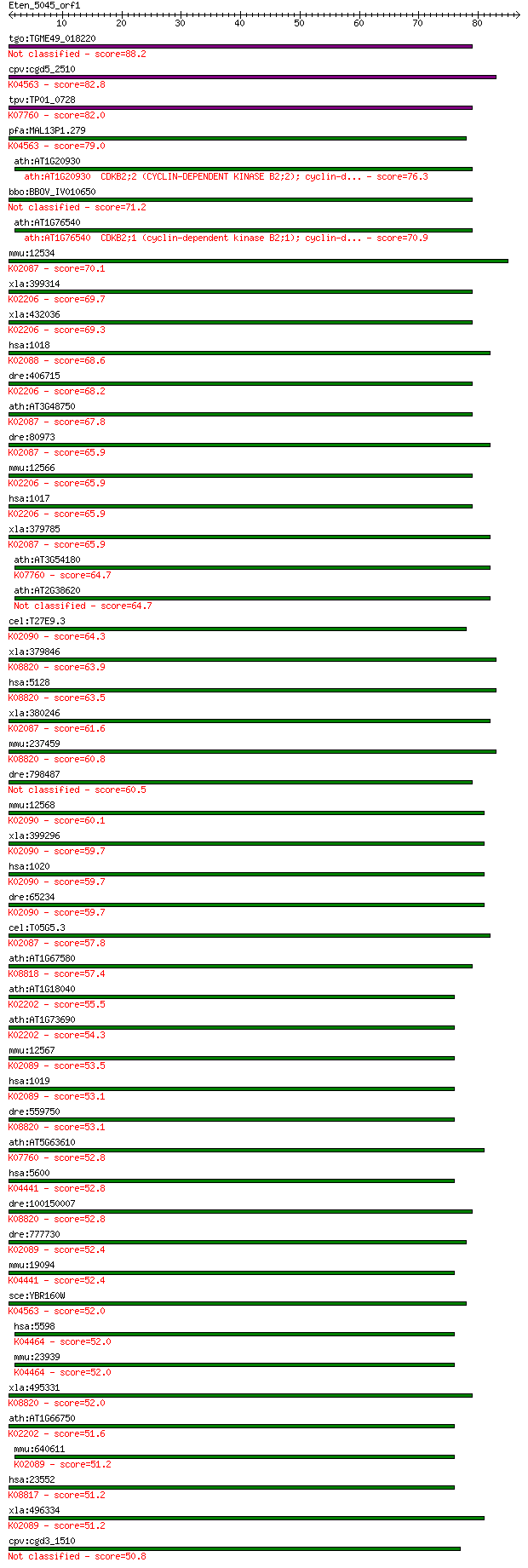
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_018220 cell division control 2-like protein kinase,... 88.2 5e-18
cpv:cgd5_2510 Cdc2-like CDK2/CDC28 like protein kinase ; K0456... 82.8 2e-16
tpv:TP01_0728 cell division control protein 2 related kinase; ... 82.0 4e-16
pfa:MAL13P1.279 PfPK5; protein kinase 5; K04563 cyclin-depende... 79.0 3e-15
ath:AT1G20930 CDKB2;2 (CYCLIN-DEPENDENT KINASE B2;2); cyclin-d... 76.3 2e-14
bbo:BBOV_IV010650 23.m06281; cell division control protein 2 (... 71.2 7e-13
ath:AT1G76540 CDKB2;1 (cyclin-dependent kinase B2;1); cyclin-d... 70.9 9e-13
mmu:12534 Cdk1, Cdc2, Cdc2a, p34; cyclin-dependent kinas... 70.1 2e-12
xla:399314 cdk2, eg1; cyclin-dependent kinase 2 (EC:2.7.11.22)... 69.7 2e-12
xla:432036 hypothetical protein MGC81499; K02206 cyclin-depend... 69.3 3e-12
hsa:1018 CDK3; cyclin-dependent kinase 3 (EC:2.7.11.22); K0208... 68.6 5e-12
dre:406715 cdk2, wu:fa10c02, zgc:56598, zgc:77684; cyclin-depe... 68.2 6e-12
ath:AT3G48750 CDC2; CDC2 (CELL DIVISION CONTROL 2); cyclin-dep... 67.8 9e-12
dre:80973 cdk1, MGC92032, cdc2, wu:fc30e01, zgc:92032; cyclin-... 65.9 3e-11
mmu:12566 Cdk2, A630093N05Rik; cyclin-dependent kinase 2 (EC:2... 65.9 3e-11
hsa:1017 CDK2, p33(CDK2); cyclin-dependent kinase 2 (EC:2.7.11... 65.9 3e-11
xla:379785 cdk1-b, MGC64249, cdc-2, cdc2, cdc2-b, cdc28a, cdc2... 65.9 3e-11
ath:AT3G54180 CDKB1;1 (CYCLIN-DEPENDENT KINASE B1;1); cyclin-d... 64.7 7e-11
ath:AT2G38620 CDKB1;2 (cyclin-dependent kinase B1;2); cyclin b... 64.7 7e-11
cel:T27E9.3 cdk-5; Cyclin-Dependent Kinase family member (cdk-... 64.3 8e-11
xla:379846 cdk17, MGC52941, PCTAIRE2, pctk2, pctk2-a; cyclin-d... 63.9 1e-10
hsa:5128 CDK17, PCTAIRE2, PCTK2; cyclin-dependent kinase 17 (E... 63.5 2e-10
xla:380246 cdk1-a, MGC53829, cdc-2, cdc2, cdc2-a, cdc28a, p34c... 61.6 5e-10
mmu:237459 Cdk17, 6430598J10Rik, MGC25109, Pctk2; cyclin-depen... 60.8 1e-09
dre:798487 cell division protein kinase 17 60.5
mmu:12568 Cdk5, AW048668, Crk6; cyclin-dependent kinase 5 (EC:... 60.1 2e-09
xla:399296 cdk5; cyclin-dependent kinase 5 (EC:2.7.11.22); K02... 59.7 2e-09
hsa:1020 CDK5, PSSALRE; cyclin-dependent kinase 5 (EC:2.7.11.2... 59.7 2e-09
dre:65234 cdk5, zgc:101604; cyclin-dependent protein kinase 5 ... 59.7 2e-09
cel:T05G5.3 cdk-1; Cyclin-Dependent Kinase family member (cdk-... 57.8 8e-09
ath:AT1G67580 protein kinase family protein; K08818 cell divis... 57.4 1e-08
ath:AT1G18040 CDKD1;3 (CYCLIN-DEPENDENT KINASE D1;3); kinase/ ... 55.5 4e-08
ath:AT1G73690 CDKD1;1 (CYCLIN-DEPENDENT KINASE D1;1); ATP bind... 54.3 9e-08
mmu:12567 Cdk4, Crk3; cyclin-dependent kinase 4 (EC:2.7.11.22)... 53.5 2e-07
hsa:1019 CDK4, CMM3, MGC14458, PSK-J3; cyclin-dependent kinase... 53.1 2e-07
dre:559750 cdk16, fc22a06, si:dkey-31k5.1, wu:fc22a06; cyclin-... 53.1 2e-07
ath:AT5G63610 CDKE;1; CDKE;1 (CYCLIN-DEPENDENT KINASE E;1); AT... 52.8 3e-07
hsa:5600 MAPK11, P38B, P38BETA2, PRKM11, SAPK2, SAPK2B, p38-2,... 52.8 3e-07
dre:100150007 PCTAIRE protein kinase 3-like; K08820 PCTAIRE pr... 52.8 3e-07
dre:777730 zgc:153726 (EC:2.7.11.22); K02089 cyclin-dependent ... 52.4 4e-07
mmu:19094 Mapk11, DKFZp586C1322, P38b, Prkm11, Sapk2, Sapk2b, ... 52.4 4e-07
sce:YBR160W CDC28, CDK1, HSL5, SRM5; Catalytic subunit of the ... 52.0 4e-07
hsa:5598 MAPK7, BMK1, ERK4, ERK5, PRKM7; mitogen-activated pro... 52.0 5e-07
mmu:23939 Mapk7, BMK1, ERK5, Erk5-T, PRKM7; mitogen-activated ... 52.0 5e-07
xla:495331 cdk16, pctaire, pctaire1, pctgaire, pctk1; cyclin-d... 52.0 5e-07
ath:AT1G66750 CAK4; CAK4 (CDK-ACTIVATING KINASE 4); kinase/ pr... 51.6 7e-07
mmu:640611 cell division protein kinase 4-like; K02089 cyclin-... 51.2 8e-07
hsa:23552 CDK20, CCRK, CDCH, P42, PNQALRE; cyclin-dependent ki... 51.2 8e-07
xla:496334 cdk6, plstire; cyclin-dependent kinase 6 (EC:2.7.11... 51.2 9e-07
cpv:cgd3_1510 cyclin-dependent kinase 3 50.8
> tgo:TGME49_018220 cell division control 2-like protein kinase,
putative (EC:2.7.11.23)
Length=300
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 42/80 (52%), Positives = 56/80 (70%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPSPTEG--LAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL +IFK+LGTP +E LA LP W +F +PP+ W +VP L G DLLS+ML F
Sbjct 208 DQLMKIFKVLGTPQVSEHPQLAELPHWNRDFPQFPPLPWDQVVPKLDPLGTDLLSRMLRF 267
Query 59 EASRRISAKTAMQHSYFDDI 78
++++RISA+ AMQH YF D+
Sbjct 268 DSNQRISARQAMQHPYFSDL 287
> cpv:cgd5_2510 Cdc2-like CDK2/CDC28 like protein kinase ; K04563
cyclin-dependent kinase [EC:2.7.11.22]
Length=295
Score = 82.8 bits (203), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 39/85 (45%), Positives = 55/85 (64%), Gaps = 3/85 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRN-NFKYYPPMKWKYIVPGLSEAGLDLLSQMLT 57
DQL +IF +LGTP+P E + LP W+ F+ + W I+PG + G+DLLS ML
Sbjct 209 DQLPKIFSILGTPNPREWPQVQELPLWKQRTFQVFEKKPWSSIIPGFCQEGIDLLSNMLC 268
Query 58 FEASRRISAKTAMQHSYFDDIDPAL 82
F+ ++RISA+ AM H YF D+DP +
Sbjct 269 FDPNKRISARDAMNHPYFKDLDPQI 293
> tpv:TP01_0728 cell division control protein 2 related kinase;
K07760 cyclin-dependent kinase [EC:2.7.11.22]
Length=298
Score = 82.0 bits (201), Expect = 4e-16, Method: Composition-based stats.
Identities = 39/80 (48%), Positives = 53/80 (66%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIFK+LGTPS + LP + +F YY W IVP L+E+G+DL+S+ML
Sbjct 208 DQLKRIFKILGTPSVDSWPQVVNLPAYNPDFSYYEKQSWSSIVPKLNESGIDLISRMLQL 267
Query 59 EASRRISAKTAMQHSYFDDI 78
+ +RISAK A++H YF D+
Sbjct 268 DPVQRISAKEALKHDYFKDL 287
> pfa:MAL13P1.279 PfPK5; protein kinase 5; K04563 cyclin-dependent
kinase [EC:2.7.11.22]
Length=288
Score = 79.0 bits (193), Expect = 3e-15, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 55/79 (69%), Gaps = 2/79 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF++LGTP+ + LP++ NF Y P+ W+ + GL E+G+DLLS+ML
Sbjct 208 DQLMRIFRILGTPNSKNWPNVTELPKYDPNFTVYEPLPWESFLKGLDESGIDLLSKMLKL 267
Query 59 EASRRISAKTAMQHSYFDD 77
+ ++RI+AK A++H+YF +
Sbjct 268 DPNQRITAKQALEHAYFKE 286
> ath:AT1G20930 CDKB2;2 (CYCLIN-DEPENDENT KINASE B2;2); cyclin-dependent
protein kinase/ kinase; K00924 [EC:2.7.1.-]
Length=315
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 40/79 (50%), Positives = 54/79 (68%), Gaps = 3/79 (3%)
Query 2 QLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFE 59
QL RIF+LLGTP+ G++ L W + + + P+ VP L EAGLDLLS+ML +E
Sbjct 232 QLLRIFRLLGTPNEEVWPGVSKLKDW-HEYPQWKPLSLSTAVPNLDEAGLDLLSKMLEYE 290
Query 60 ASRRISAKTAMQHSYFDDI 78
++RISAK AM+H YFDD+
Sbjct 291 PAKRISAKKAMEHPYFDDL 309
> bbo:BBOV_IV010650 23.m06281; cell division control protein 2
(EC:2.7.11.1)
Length=295
Score = 71.2 bits (173), Expect = 7e-13, Method: Composition-based stats.
Identities = 34/80 (42%), Positives = 50/80 (62%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPS--PTEGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIFK+LG+P+ G+ LP + + + W IVP L AG+DL+S+ML
Sbjct 208 DQLKRIFKVLGSPNVGTWPGVVDLPAYNPDMDQFEKQPWNVIVPKLGGAGVDLISKMLQL 267
Query 59 EASRRISAKTAMQHSYFDDI 78
+ +RISA+ A+ H YF+D+
Sbjct 268 DPFQRISARDALCHEYFNDV 287
> ath:AT1G76540 CDKB2;1 (cyclin-dependent kinase B2;1); cyclin-dependent
protein kinase/ kinase/ protein binding; K00924
[EC:2.7.1.-]
Length=313
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 38/79 (48%), Positives = 51/79 (64%), Gaps = 3/79 (3%)
Query 2 QLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFE 59
QL IFKL GTP+ G++ L W + + + P VP L EAG+DLLS+ML +E
Sbjct 230 QLLHIFKLFGTPNEEMWPGVSTLKNW-HEYPQWKPSTLSSAVPNLDEAGVDLLSKMLQYE 288
Query 60 ASRRISAKTAMQHSYFDDI 78
++RISAK AM+H YFDD+
Sbjct 289 PAKRISAKMAMEHPYFDDL 307
> mmu:12534 Cdk1, Cdc2, Cdc2a, p34; cyclin-dependent kinase
1 (EC:2.7.11.23 2.7.11.22); K02087 cyclin-dependent kinase
1 [EC:2.7.11.22]
Length=297
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 36/86 (41%), Positives = 53/86 (61%), Gaps = 2/86 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP+ + L ++N F + P V L E GLDLLS+ML +
Sbjct 211 DQLFRIFRALGTPNNEVWPEVESLQDYKNTFPKWKPGSLASHVKNLDENGLDLLSKMLVY 270
Query 59 EASRRISAKTAMQHSYFDDIDPALRE 84
+ ++RIS K A++H YFDD+D +++
Sbjct 271 DPAKRISGKMALKHPYFDDLDNQIKK 296
> xla:399314 cdk2, eg1; cyclin-dependent kinase 2 (EC:2.7.11.22);
K02206 cyclin-dependent kinase 2 [EC:2.7.11.22]
Length=297
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 51/80 (63%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP G+ +P +++ F + + +VP L E G DLL+QML +
Sbjct 210 DQLFRIFRTLGTPDEVSWPGVTTMPDYKSTFPKWIRQDFSKVVPPLDEDGRDLLAQMLQY 269
Query 59 EASRRISAKTAMQHSYFDDI 78
++++RISAK A+ H +F D+
Sbjct 270 DSNKRISAKVALTHPFFRDV 289
> xla:432036 hypothetical protein MGC81499; K02206 cyclin-dependent
kinase 2 [EC:2.7.11.22]
Length=297
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 51/80 (63%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP G+ +P +++ F + + +VP L E G DLL+QML +
Sbjct 210 DQLFRIFRTLGTPDEVSWPGVTTMPDYKSTFPKWIRQDFSKVVPPLDEDGRDLLAQMLQY 269
Query 59 EASRRISAKTAMQHSYFDDI 78
++++RISAK A+ H +F D+
Sbjct 270 DSNKRISAKAALTHPFFRDV 289
> hsa:1018 CDK3; cyclin-dependent kinase 3 (EC:2.7.11.22); K02088
cyclin-dependent kinase 3 [EC:2.7.11.22]
Length=305
Score = 68.6 bits (166), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 52/83 (62%), Gaps = 2/83 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF++LGTPS G+ LP ++ +F + + IVP L G DLL Q+L +
Sbjct 210 DQLFRIFRMLGTPSEDTWPGVTQLPDYKGSFPKWTRKGLEEIVPNLEPEGRDLLMQLLQY 269
Query 59 EASRRISAKTAMQHSYFDDIDPA 81
+ S+RI+AKTA+ H YF +P+
Sbjct 270 DPSQRITAKTALAHPYFSSPEPS 292
> dre:406715 cdk2, wu:fa10c02, zgc:56598, zgc:77684; cyclin-dependent
kinase 2 (EC:2.7.1.-); K02206 cyclin-dependent kinase
2 [EC:2.7.11.22]
Length=298
Score = 68.2 bits (165), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 34/80 (42%), Positives = 50/80 (62%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPSPT--EGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP + G+ +P ++ +F + +VP L E G DLL QMLT+
Sbjct 210 DQLFRIFRTLGTPDESIWPGVTSMPDYKPSFPKWARQDLSKVVPPLDEDGRDLLGQMLTY 269
Query 59 EASRRISAKTAMQHSYFDDI 78
+ ++RISAK A+ H +F D+
Sbjct 270 DPNKRISAKNALVHRFFRDV 289
> ath:AT3G48750 CDC2; CDC2 (CELL DIVISION CONTROL 2); cyclin-dependent
protein kinase/ kinase/ protein binding / protein kinase;
K02087 cyclin-dependent kinase 1 [EC:2.7.11.22]
Length=294
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 31/80 (38%), Positives = 51/80 (63%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPSPT--EGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL +IF+++GTP G+ LP +++ F + P + VP L G+DLLS+ML
Sbjct 211 DQLFKIFRIMGTPYEDTWRGVTSLPDYKSAFPKWKPTDLETFVPNLDPDGVDLLSKMLLM 270
Query 59 EASRRISAKTAMQHSYFDDI 78
+ ++RI+A+ A++H YF D+
Sbjct 271 DPTKRINARAALEHEYFKDL 290
> dre:80973 cdk1, MGC92032, cdc2, wu:fc30e01, zgc:92032; cyclin-dependent
kinase 1; K02087 cyclin-dependent kinase 1 [EC:2.7.11.22]
Length=302
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 49/83 (59%), Gaps = 2/83 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP+ + LP ++N F + V L + G+DLL +ML +
Sbjct 211 DQLFRIFRTLGTPNNEVWPDVESLPDYKNTFPKWKSGNLANTVKNLDKNGIDLLMKMLIY 270
Query 59 EASRRISAKTAMQHSYFDDIDPA 81
+ +RISA+ AM H YFDD+D +
Sbjct 271 DPPKRISARQAMTHPYFDDLDKS 293
> mmu:12566 Cdk2, A630093N05Rik; cyclin-dependent kinase 2 (EC:2.7.11.22);
K02206 cyclin-dependent kinase 2 [EC:2.7.11.22]
Length=298
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 49/80 (61%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP G+ +P ++ +F + + +VP L E G LLSQML +
Sbjct 210 DQLFRIFRTLGTPDEVVWPGVTSMPDYKPSFPKWARQDFSKVVPPLDEDGRSLLSQMLHY 269
Query 59 EASRRISAKTAMQHSYFDDI 78
+ ++RISAK A+ H +F D+
Sbjct 270 DPNKRISAKAALAHPFFQDV 289
> hsa:1017 CDK2, p33(CDK2); cyclin-dependent kinase 2 (EC:2.7.11.22);
K02206 cyclin-dependent kinase 2 [EC:2.7.11.22]
Length=298
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 49/80 (61%), Gaps = 2/80 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP G+ +P ++ +F + + +VP L E G LLSQML +
Sbjct 210 DQLFRIFRTLGTPDEVVWPGVTSMPDYKPSFPKWARQDFSKVVPPLDEDGRSLLSQMLHY 269
Query 59 EASRRISAKTAMQHSYFDDI 78
+ ++RISAK A+ H +F D+
Sbjct 270 DPNKRISAKAALAHPFFQDV 289
> xla:379785 cdk1-b, MGC64249, cdc-2, cdc2, cdc2-b, cdc28a, cdc2a,
cdc2x1.2, p34cdc2, xcdc2; cyclin-dependent kinase 1 (EC:2.7.11.23
2.7.11.22); K02087 cyclin-dependent kinase 1 [EC:2.7.11.22]
Length=302
Score = 65.9 bits (159), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 50/83 (60%), Gaps = 2/83 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP+ + L ++N F + V + E GLDLLS+ML +
Sbjct 211 DQLFRIFRSLGTPNNEVWPEVESLQDYKNTFPKWKGGSLSSNVKNIDEDGLDLLSKMLVY 270
Query 59 EASRRISAKTAMQHSYFDDIDPA 81
+ ++RISA+ AM H YFDD+D +
Sbjct 271 DPAKRISARKAMLHPYFDDLDKS 293
> ath:AT3G54180 CDKB1;1 (CYCLIN-DEPENDENT KINASE B1;1); cyclin-dependent
protein kinase/ kinase/ protein binding; K07760 cyclin-dependent
kinase [EC:2.7.11.22]
Length=309
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 35/82 (42%), Positives = 51/82 (62%), Gaps = 3/82 (3%)
Query 2 QLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFE 59
QL IF+LLGTP+ + G++ L W + + + P VP LS G+DLL++ML +
Sbjct 227 QLLHIFRLLGTPTEQQWPGVSTLRDW-HVYPKWEPQDLTLAVPSLSPQGVDLLTKMLKYN 285
Query 60 ASRRISAKTAMQHSYFDDIDPA 81
+ RISAKTA+ H YFD +D +
Sbjct 286 PAERISAKTALDHPYFDSLDKS 307
> ath:AT2G38620 CDKB1;2 (cyclin-dependent kinase B1;2); cyclin
binding / kinase/ protein serine/threonine kinase
Length=311
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 35/82 (42%), Positives = 49/82 (59%), Gaps = 3/82 (3%)
Query 2 QLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFE 59
QL IF+LLGTP+ + G+ L W + + + P VP LS G+DLL+QML +
Sbjct 229 QLLHIFRLLGTPTEQQWPGVMALRDW-HVYPKWEPQDLSRAVPSLSPEGIDLLTQMLKYN 287
Query 60 ASRRISAKTAMQHSYFDDIDPA 81
+ RISAK A+ H YFD +D +
Sbjct 288 PAERISAKAALDHPYFDSLDKS 309
> cel:T27E9.3 cdk-5; Cyclin-Dependent Kinase family member (cdk-5);
K02090 cyclin-dependent kinase 5 [EC:2.7.11.22]
Length=292
Score = 64.3 bits (155), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 46/79 (58%), Gaps = 2/79 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIFK LG+PS + LP ++ Y+P + W IVP L+ G DLL ++L
Sbjct 210 DQLKRIFKQLGSPSEDNWPSITQLPDYKPYPIYHPTLTWSQIVPNLNSRGRDLLQKLLVC 269
Query 59 EASRRISAKTAMQHSYFDD 77
+ RI A A++H+YF D
Sbjct 270 NPAGRIDADAALRHAYFAD 288
> xla:379846 cdk17, MGC52941, PCTAIRE2, pctk2, pctk2-a; cyclin-dependent
kinase 17 (EC:2.7.11.22); K08820 PCTAIRE protein
kinase [EC:2.7.11.22]
Length=500
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 52/85 (61%), Gaps = 3/85 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRN-NFKYYPPMKWKYIVPGLSEAGLDLLSQMLT 57
D+LH IF+LLGTP+ G++ ++RN NF Y P P L G++LL++ L
Sbjct 373 DELHLIFRLLGTPAEETWPGISSNDEFRNYNFPKYKPQPLINHAPRLDSEGIELLTRFLQ 432
Query 58 FEASRRISAKTAMQHSYFDDIDPAL 82
+E+ +RISA+ AM+H+YF + L
Sbjct 433 YESKKRISAEDAMKHAYFRSLGTKL 457
> hsa:5128 CDK17, PCTAIRE2, PCTK2; cyclin-dependent kinase 17
(EC:2.7.11.22); K08820 PCTAIRE protein kinase [EC:2.7.11.22]
Length=523
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 32/85 (37%), Positives = 52/85 (61%), Gaps = 3/85 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRN-NFKYYPPMKWKYIVPGLSEAGLDLLSQMLT 57
D+LH IF+LLGTPS G++ +++N NF Y P P L G++L+++ L
Sbjct 396 DELHLIFRLLGTPSQETWPGISSNEEFKNYNFPKYKPQPLINHAPRLDSEGIELITKFLQ 455
Query 58 FEASRRISAKTAMQHSYFDDIDPAL 82
+E+ +R+SA+ AM+H YF + P +
Sbjct 456 YESKKRVSAEEAMKHVYFRSLGPRI 480
> xla:380246 cdk1-a, MGC53829, cdc-2, cdc2, cdc2-a, cdc28a, p34cdc2,
xcdc2; cyclin-dependent kinase 1 (EC:2.7.11.23 2.7.11.22);
K02087 cyclin-dependent kinase 1 [EC:2.7.11.22]
Length=302
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 32/83 (38%), Positives = 51/83 (61%), Gaps = 2/83 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+ LGTP+ + L ++N+F + V + + GLDLL++ML +
Sbjct 211 DQLFRIFRALGTPNNEVWPEVESLQDYKNSFPKWKGGSLSANVKNIDKDGLDLLAKMLIY 270
Query 59 EASRRISAKTAMQHSYFDDIDPA 81
+ ++RISA+ A+ H YFDD+D +
Sbjct 271 DPAKRISARKALLHPYFDDLDKS 293
> mmu:237459 Cdk17, 6430598J10Rik, MGC25109, Pctk2; cyclin-dependent
kinase 17 (EC:2.7.11.22); K08820 PCTAIRE protein kinase
[EC:2.7.11.22]
Length=523
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/85 (36%), Positives = 51/85 (60%), Gaps = 3/85 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRN-NFKYYPPMKWKYIVPGLSEAGLDLLSQMLT 57
D+LH IF+LLGTPS G++ +++N NF Y P P L G++L+++ L
Sbjct 396 DELHLIFRLLGTPSQETWPGVSSNDEFKNYNFPKYKPQPLINHAPRLDSEGIELITKFLQ 455
Query 58 FEASRRISAKTAMQHSYFDDIDPAL 82
+E+ +R+ A+ AM+H YF + P +
Sbjct 456 YESKKRVPAEEAMKHVYFRSLGPRI 480
> dre:798487 cell division protein kinase 17
Length=526
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 50/81 (61%), Gaps = 3/81 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRN-NFKYYPPMKWKYIVPGLSEAGLDLLSQMLT 57
D+LH IF+LLGTP+ G++ + ++++ NF Y P + P L G++LL L
Sbjct 399 DELHLIFRLLGTPTEDNWPGISSIEEFKSYNFPKYKPQPFINHAPRLDTEGIELLLSFLR 458
Query 58 FEASRRISAKTAMQHSYFDDI 78
+E+ +RISA +M+HSYF +
Sbjct 459 YESKKRISADESMKHSYFKSL 479
> mmu:12568 Cdk5, AW048668, Crk6; cyclin-dependent kinase 5 (EC:2.7.11.22);
K02090 cyclin-dependent kinase 5 [EC:2.7.11.22]
Length=292
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 33/82 (40%), Positives = 45/82 (54%), Gaps = 2/82 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+LLGTP+ + + LP ++ Y +VP L+ G DLL +L
Sbjct 210 DQLKRIFRLLGTPTEEQWPAMTKLPDYKPYPMYPATTSLVNVVPKLNATGRDLLQNLLKC 269
Query 59 EASRRISAKTAMQHSYFDDIDP 80
+RISA+ A+QH YF D P
Sbjct 270 NPVQRISAEEALQHPYFSDFCP 291
> xla:399296 cdk5; cyclin-dependent kinase 5 (EC:2.7.11.22); K02090
cyclin-dependent kinase 5 [EC:2.7.11.22]
Length=292
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 33/82 (40%), Positives = 44/82 (53%), Gaps = 2/82 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+LLGTP+ + + LP ++ Y M +VP L+ G DLL +L
Sbjct 210 DQLKRIFRLLGTPTEEQWPAMTKLPDYKPYPMYPATMSLVNVVPKLNATGRDLLQNLLKC 269
Query 59 EASRRISAKTAMQHSYFDDIDP 80
+RI A A+QH YF D P
Sbjct 270 NPVQRICADEALQHPYFADFCP 291
> hsa:1020 CDK5, PSSALRE; cyclin-dependent kinase 5 (EC:2.7.11.22);
K02090 cyclin-dependent kinase 5 [EC:2.7.11.22]
Length=260
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 33/82 (40%), Positives = 45/82 (54%), Gaps = 2/82 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+LLGTP+ + + LP ++ Y +VP L+ G DLL +L
Sbjct 178 DQLKRIFRLLGTPTEEQWPSMTKLPDYKPYPMYPATTSLVNVVPKLNATGRDLLQNLLKC 237
Query 59 EASRRISAKTAMQHSYFDDIDP 80
+RISA+ A+QH YF D P
Sbjct 238 NPVQRISAEEALQHPYFSDFCP 259
> dre:65234 cdk5, zgc:101604; cyclin-dependent protein kinase
5 (EC:2.7.11.22); K02090 cyclin-dependent kinase 5 [EC:2.7.11.22]
Length=292
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 34/82 (41%), Positives = 45/82 (54%), Gaps = 2/82 (2%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL RIF+LLGTP+ + + LP ++ Y +VP LS G DLL +L
Sbjct 210 DQLKRIFRLLGTPTEEQWQTMNKLPDYKPYPMYPATTSLVNVVPKLSSTGRDLLQNLLKC 269
Query 59 EASRRISAKTAMQHSYFDDIDP 80
+RISA+ A+QH YF D P
Sbjct 270 NPVQRISAEEALQHPYFADFCP 291
> cel:T05G5.3 cdk-1; Cyclin-Dependent Kinase family member (cdk-1);
K02087 cyclin-dependent kinase 1 [EC:2.7.11.22]
Length=332
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 34/93 (36%), Positives = 53/93 (56%), Gaps = 15/93 (16%)
Query 1 DQLHRIFKLLGTPSPTE--GL-------AGLPQWRNNF---KYYPPMKWKYIVPGLSEAG 48
D+L RIF++LGTP+ E G+ A P+WR NF K+Y K++ L +
Sbjct 229 DELFRIFRVLGTPTELEWNGVESLPDYKATFPKWRENFLRDKFYDKKTGKHL---LDDTA 285
Query 49 LDLLSQMLTFEASRRISAKTAMQHSYFDDIDPA 81
LL +L ++ S R++AK A+ H YFD++D +
Sbjct 286 FSLLEGLLIYDPSLRLNAKKALVHPYFDNMDTS 318
> ath:AT1G67580 protein kinase family protein; K08818 cell division
cycle 2-like [EC:2.7.11.22]
Length=752
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 32/89 (35%), Positives = 53/89 (59%), Gaps = 12/89 (13%)
Query 1 DQLHRIFKLLGTPSPT--EGLAGLPQWRNNF---------KYYPPMKWKYIVPGLSEAGL 49
DQL +IF++LGTP+ + G + LP + NF K +P + P LS+AG
Sbjct 613 DQLDKIFRILGTPNESIWPGFSKLPGVKVNFVKHQYNLLRKKFPATSFTG-APVLSDAGF 671
Query 50 DLLSQMLTFEASRRISAKTAMQHSYFDDI 78
DLL+++LT++ RRI+ A++H +F ++
Sbjct 672 DLLNKLLTYDPERRITVNEALKHDWFREV 700
> ath:AT1G18040 CDKD1;3 (CYCLIN-DEPENDENT KINASE D1;3); kinase/
protein kinase; K02202 cyclin-dependent kinase 7 [EC:2.7.11.22]
Length=391
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 44/77 (57%), Gaps = 3/77 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL +IF GTP + L LP + +++ P + + P +S+ LDLLS+M T+
Sbjct 217 DQLSKIFAAFGTPKADQWPDLTKLPDY-VEYQFVPAPSLRSLFPAVSDDALDLLSKMFTY 275
Query 59 EASRRISAKTAMQHSYF 75
+ RIS K A++H YF
Sbjct 276 DPKARISIKQALEHRYF 292
> ath:AT1G73690 CDKD1;1 (CYCLIN-DEPENDENT KINASE D1;1); ATP binding
/ kinase/ protein kinase/ protein serine/threonine kinase;
K02202 cyclin-dependent kinase 7 [EC:2.7.11.22]
Length=398
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 45/77 (58%), Gaps = 3/77 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL +IF GTP + + LP + +++ P + ++P +SE LDLLS+M T+
Sbjct 216 DQLSKIFAAFGTPKADQWPDMICLPDY-VEYQFVPAPSLRSLLPTVSEDALDLLSKMFTY 274
Query 59 EASRRISAKTAMQHSYF 75
+ RIS + A++H YF
Sbjct 275 DPKSRISIQQALKHRYF 291
> mmu:12567 Cdk4, Crk3; cyclin-dependent kinase 4 (EC:2.7.11.22);
K02089 cyclin-dependent kinase 4 [EC:2.7.11.22]
Length=303
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 30/75 (40%), Positives = 41/75 (54%), Gaps = 0/75 (0%)
Query 1 DQLHRIFKLLGTPSPTEGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFEA 60
DQL +IF L+G P + + R F P + +VP + E+G LL +MLTF
Sbjct 221 DQLGKIFDLIGLPPEDDWPREVSLPRGAFAPRGPRPVQSVVPEMEESGAQLLLEMLTFNP 280
Query 61 SRRISAKTAMQHSYF 75
+RISA A+QHSY
Sbjct 281 HKRISAFRALQHSYL 295
> hsa:1019 CDK4, CMM3, MGC14458, PSK-J3; cyclin-dependent kinase
4 (EC:2.7.11.22); K02089 cyclin-dependent kinase 4 [EC:2.7.11.22]
Length=303
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 30/75 (40%), Positives = 41/75 (54%), Gaps = 0/75 (0%)
Query 1 DQLHRIFKLLGTPSPTEGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFEA 60
DQL +IF L+G P + + R F P + +VP + E+G LL +MLTF
Sbjct 221 DQLGKIFDLIGLPPEDDWPRDVSLPRGAFPPRGPRPVQSVVPEMEESGAQLLLEMLTFNP 280
Query 61 SRRISAKTAMQHSYF 75
+RISA A+QHSY
Sbjct 281 HKRISAFRALQHSYL 295
> dre:559750 cdk16, fc22a06, si:dkey-31k5.1, wu:fc22a06; cyclin-dependent
kinase 16; K08820 PCTAIRE protein kinase [EC:2.7.11.22]
Length=524
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/79 (40%), Positives = 46/79 (58%), Gaps = 5/79 (6%)
Query 1 DQLHRIFKLLGTPSPTEGLAGLPQWRNNFKY-YPPMKWKYI---VPGLSEAGLDLLSQML 56
++LH IFKLLGTP+ E G+ Y YP + + P L G++LLS++L
Sbjct 397 EELHFIFKLLGTPTE-ETWPGITSNEEFISYNYPRYRADCLHNHTPRLDNDGVELLSKLL 455
Query 57 TFEASRRISAKTAMQHSYF 75
FE +RI+A+ AM+H YF
Sbjct 456 QFEGKKRIAAEEAMRHPYF 474
> ath:AT5G63610 CDKE;1; CDKE;1 (CYCLIN-DEPENDENT KINASE E;1);
ATP binding / kinase/ protein kinase/ protein serine/threonine
kinase; K07760 cyclin-dependent kinase [EC:2.7.11.22]
Length=470
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 53/90 (58%), Gaps = 13/90 (14%)
Query 1 DQLHRIFKLLGTPS----PTEGLAGLPQWRNNFKYYPPMKWKYI----VPGLSEA--GLD 50
DQL +IFK+LG P+ PT L LP W+N+ ++ K+ + V L++ D
Sbjct 251 DQLDKIFKILGHPTMDKWPT--LVNLPHWQNDVQHIQAHKYDSVGLHNVVHLNQKSPAYD 308
Query 51 LLSQMLTFEASRRISAKTAMQHSYFDDIDP 80
LLS+ML ++ +RI+A A++H YF +DP
Sbjct 309 LLSKMLEYDPLKRITASQALEHEYF-RMDP 337
> hsa:5600 MAPK11, P38B, P38BETA2, PRKM11, SAPK2, SAPK2B, p38-2,
p38Beta; mitogen-activated protein kinase 11 (EC:2.7.11.24);
K04441 p38 MAP kinase [EC:2.7.11.24]
Length=364
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 46/80 (57%), Gaps = 6/80 (7%)
Query 1 DQLHRIFKLLGTPSPTEGLAGLP--QWRNNFKYYPPMKWK---YIVPGLSEAGLDLLSQM 55
DQL RI +++GTPSP E LA + R + PPM K I G + +DLL +M
Sbjct 230 DQLKRIMEVVGTPSP-EVLAKISSEHARTYIQSLPPMPQKDLSSIFRGANPLAIDLLGRM 288
Query 56 LTFEASRRISAKTAMQHSYF 75
L ++ +R+SA A+ H+YF
Sbjct 289 LVLDSDQRVSAAEALAHAYF 308
> dre:100150007 PCTAIRE protein kinase 3-like; K08820 PCTAIRE
protein kinase [EC:2.7.11.22]
Length=470
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 48/81 (59%), Gaps = 3/81 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNN-FKYYPPMKWKYIVPGLSEAGLDLLSQMLT 57
++LH IF+L+GTP+ G+ ++++ F Y VP L G+DLL+ +L
Sbjct 337 EELHLIFRLMGTPTEESWPGITANEEFKSYLFPQYRAQALINHVPRLDTEGIDLLTALLL 396
Query 58 FEASRRISAKTAMQHSYFDDI 78
++ RRISA+ +++HSYF +
Sbjct 397 YDTKRRISAELSLRHSYFQTL 417
> dre:777730 zgc:153726 (EC:2.7.11.22); K02089 cyclin-dependent
kinase 4 [EC:2.7.11.22]
Length=297
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 43/77 (55%), Gaps = 0/77 (0%)
Query 1 DQLHRIFKLLGTPSPTEGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFEA 60
DQL +IF ++G P+ + + +NF P VP ++E G +LL +MLTF+
Sbjct 221 DQLGKIFAVIGLPAEDQWPTDVTLSHHNFSPQSPRPITDCVPDITEKGAELLLKMLTFDP 280
Query 61 SRRISAKTAMQHSYFDD 77
+RISA A+ H +F +
Sbjct 281 LKRISALNALDHPFFSE 297
> mmu:19094 Mapk11, DKFZp586C1322, P38b, Prkm11, Sapk2, Sapk2b,
p38-2, p38beta, p38beta2; mitogen-activated protein kinase
11 (EC:2.7.11.24); K04441 p38 MAP kinase [EC:2.7.11.24]
Length=364
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 31/80 (38%), Positives = 46/80 (57%), Gaps = 6/80 (7%)
Query 1 DQLHRIFKLLGTPSPTEGLAGLP--QWRNNFKYYPPMKWK---YIVPGLSEAGLDLLSQM 55
DQL RI +++GTPSP E LA + R + PPM K + G + +DLL +M
Sbjct 230 DQLKRIMEVVGTPSP-EVLAKISSEHARTYIQSLPPMPQKDLSSVFHGANPLAIDLLGRM 288
Query 56 LTFEASRRISAKTAMQHSYF 75
L ++ +R+SA A+ H+YF
Sbjct 289 LVLDSDQRVSAAEALAHAYF 308
> sce:YBR160W CDC28, CDK1, HSL5, SRM5; Catalytic subunit of the
main cell cycle cyclin-dependent kinase (CDK); alternately
associates with G1 cyclins (CLNs) and G2/M cyclins (CLBs) which
direct the CDK to specific substrates (EC:2.7.11.22); K04563
cyclin-dependent kinase [EC:2.7.11.22]
Length=298
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 45/79 (56%), Gaps = 2/79 (2%)
Query 1 DQLHRIFKLLGTPSPT--EGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQ+ +IF++LGTP+ + LP ++ +F + +VP L G+DLL ++L +
Sbjct 219 DQIFKIFRVLGTPNEAIWPDIVYLPDFKPSFPQWRRKDLSQVVPSLDPRGIDLLDKLLAY 278
Query 59 EASRRISAKTAMQHSYFDD 77
+ RISA+ A H YF +
Sbjct 279 DPINRISARRAAIHPYFQE 297
> hsa:5598 MAPK7, BMK1, ERK4, ERK5, PRKM7; mitogen-activated protein
kinase 7 (EC:2.7.11.24); K04464 mitogen-activated protein
kinase 7 [EC:2.7.11.24]
Length=816
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 41/78 (52%), Gaps = 4/78 (5%)
Query 2 QLHRIFKLLGTPSPTEGLA-GLPQWRNNFKYYPP---MKWKYIVPGLSEAGLDLLSQMLT 57
QL I +LGTPSP A G + R + PP + W+ + PG L LL +ML
Sbjct 270 QLQLIMMVLGTPSPAVIQAVGAERVRAYIQSLPPRQPVPWETVYPGADRQALSLLGRMLR 329
Query 58 FEASRRISAKTAMQHSYF 75
FE S RISA A++H +
Sbjct 330 FEPSARISAAAALRHPFL 347
> mmu:23939 Mapk7, BMK1, ERK5, Erk5-T, PRKM7; mitogen-activated
protein kinase 7 (EC:2.7.11.24); K04464 mitogen-activated
protein kinase 7 [EC:2.7.11.24]
Length=806
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 31/78 (39%), Positives = 41/78 (52%), Gaps = 4/78 (5%)
Query 2 QLHRIFKLLGTPSPTEGLA-GLPQWRNNFKYYPP---MKWKYIVPGLSEAGLDLLSQMLT 57
QL I +LGTPSP A G + R + PP + W+ + PG L LL +ML
Sbjct 270 QLQLIMMVLGTPSPAVIQAVGAERVRAYIQSLPPRQPVPWETVYPGADRQALSLLGRMLR 329
Query 58 FEASRRISAKTAMQHSYF 75
FE S RISA A++H +
Sbjct 330 FEPSARISAAAALRHPFL 347
> xla:495331 cdk16, pctaire, pctaire1, pctgaire, pctk1; cyclin-dependent
kinase 16 (EC:2.7.11.22); K08820 PCTAIRE protein
kinase [EC:2.7.11.22]
Length=522
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/82 (39%), Positives = 48/82 (58%), Gaps = 5/82 (6%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWR--NNFKYYPPMKWKYIVPGLSEAGLDLLSQML 56
+QLH IF++LGTP+ G+ +++ N KYYP K+ L G LLS++L
Sbjct 396 EQLHFIFRILGTPTEETWPGILSNEEFKSYNYPKYYPDPIQKH-AARLDSDGAKLLSKLL 454
Query 57 TFEASRRISAKTAMQHSYFDDI 78
E +RISA+ AM+H YF ++
Sbjct 455 QLEGRKRISAEEAMKHLYFQEL 476
> ath:AT1G66750 CAK4; CAK4 (CDK-ACTIVATING KINASE 4); kinase/
protein binding / protein serine/threonine kinase; K02202 cyclin-dependent
kinase 7 [EC:2.7.11.22]
Length=348
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 44/77 (57%), Gaps = 3/77 (3%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTF 58
DQL +IF+ GTP P++ + LP + F Y P + I P S+ LDLL++M +
Sbjct 218 DQLGKIFQAFGTPVPSQWSDMIYLPDYME-FSYTPAPPLRTIFPMASDDALDLLAKMFIY 276
Query 59 EASRRISAKTAMQHSYF 75
+ +RI+ + A+ H YF
Sbjct 277 DPRQRITIQQALDHRYF 293
> mmu:640611 cell division protein kinase 4-like; K02089 cyclin-dependent
kinase 4 [EC:2.7.11.22]
Length=303
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 29/74 (39%), Positives = 40/74 (54%), Gaps = 0/74 (0%)
Query 2 QLHRIFKLLGTPSPTEGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFEAS 61
QL +IF L+G P + + R F P + +VP + E+G LL +MLTF
Sbjct 222 QLGKIFDLIGLPPEDDWPREVSLPRGAFAPRGPRPVQSVVPEMEESGAQLLLEMLTFNPH 281
Query 62 RRISAKTAMQHSYF 75
+RISA A+QHSY
Sbjct 282 KRISAFRALQHSYL 295
> hsa:23552 CDK20, CCRK, CDCH, P42, PNQALRE; cyclin-dependent
kinase 20 (EC:2.7.11.22); K08817 cell cycle related kinase [EC:2.7.11.22]
Length=346
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 29/78 (37%), Positives = 44/78 (56%), Gaps = 3/78 (3%)
Query 1 DQLHRIFKLLGTPSPT--EGLAGLPQWRN-NFKYYPPMKWKYIVPGLSEAGLDLLSQMLT 57
+QL + ++LGTP+P L LP + +FK PM + ++P +S LDLL Q L
Sbjct 211 EQLCYVLRILGTPNPQVWPELTELPDYNKISFKEQVPMPLEEVLPDVSPQALDLLGQFLL 270
Query 58 FEASRRISAKTAMQHSYF 75
+ +RI+A A+ H YF
Sbjct 271 YPPHQRIAASKALLHQYF 288
> xla:496334 cdk6, plstire; cyclin-dependent kinase 6 (EC:2.7.11.22);
K02089 cyclin-dependent kinase 4 [EC:2.7.11.22]
Length=319
Score = 51.2 bits (121), Expect = 9e-07, Method: Composition-based stats.
Identities = 30/80 (37%), Positives = 43/80 (53%), Gaps = 1/80 (1%)
Query 1 DQLHRIFKLLGTPSPTEGLAGLPQWRNNFKYYPPMKWKYIVPGLSEAGLDLLSQMLTFEA 60
DQL +IF+++G PS E A + R+ F + VP + G DLL MLTF
Sbjct 221 DQLCKIFEMIGLPSEEEWPADVTLSRSAFSPRTQQPVEKFVPEIDPNGADLLLAMLTFSP 280
Query 61 SRRISAKTAMQHSYFDDIDP 80
+R+SA A+ H +F + DP
Sbjct 281 QKRVSASDALLHPFFAE-DP 299
> cpv:cgd3_1510 cyclin-dependent kinase 3
Length=331
Score = 50.8 bits (120), Expect = 9e-07, Method: Composition-based stats.
Identities = 28/81 (34%), Positives = 48/81 (59%), Gaps = 5/81 (6%)
Query 1 DQLHRIFKLLGTPSPTE--GLAGLPQWRNNF---KYYPPMKWKYIVPGLSEAGLDLLSQM 55
D L IF+LLGT + + G+ LP +++ F K P + ++P L +AG+DLL ++
Sbjct 239 DTLFYIFRLLGTANESNWPGVTQLPCYKSVFPQWKVNPKLNLHALLPNLDQAGVDLLFRL 298
Query 56 LTFEASRRISAKTAMQHSYFD 76
L + +RI+A A+QH + +
Sbjct 299 LQYCPKKRITALEALQHPWLN 319
Lambda K H
0.319 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2030857360
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40