bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4999_orf1
Length=138
Score E
Sequences producing significant alignments: (Bits) Value
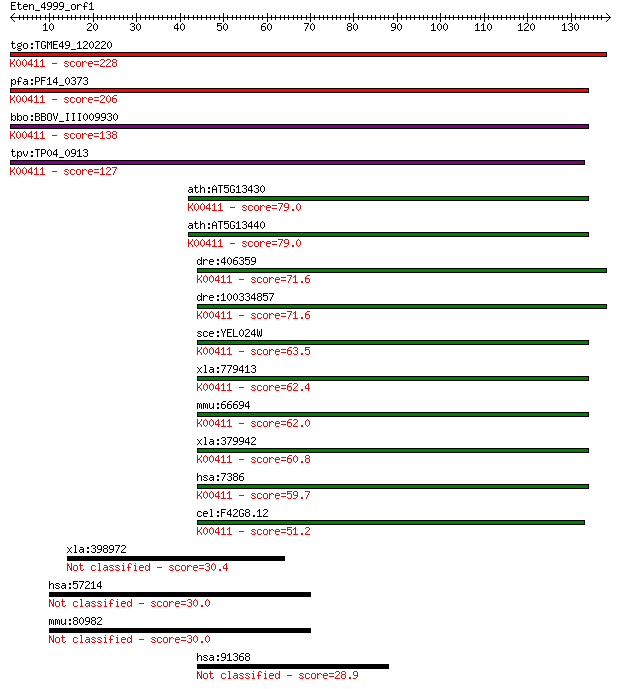
tgo:TGME49_120220 ubiquinol-cytochrome c reductase domain-cont... 228 4e-60
pfa:PF14_0373 ubiquinol-cytochrome c reductase iron-sulfur sub... 206 1e-53
bbo:BBOV_III009930 17.m07861; ubiquinol cytochrome c oxidoredu... 138 6e-33
tpv:TP04_0913 ubiquinol-cytochrome C reductase, iron-sulfur su... 127 1e-29
ath:AT5G13430 ubiquinol-cytochrome C reductase iron-sulfur sub... 79.0 4e-15
ath:AT5G13440 ubiquinol-cytochrome C reductase iron-sulfur sub... 79.0 5e-15
dre:406359 uqcrfs1, MGC174806, fb78f10, wu:fb05d11, wu:fb78f10... 71.6 8e-13
dre:100334857 ubiquinol-cytochrome c reductase, Rieske iron-su... 71.6 8e-13
sce:YEL024W RIP1; Rip1p (EC:1.10.2.2); K00411 ubiquinol-cytoch... 63.5 2e-10
xla:779413 hypothetical protein MGC154858; K00411 ubiquinol-cy... 62.4 4e-10
mmu:66694 Uqcrfs1, 4430402G14Rik, AI875505; ubiquinol-cytochro... 62.0 5e-10
xla:379942 uqcrfs1, MGC53134; ubiquinol-cytochrome c reductase... 60.8 1e-09
hsa:7386 UQCRFS1, RIP1, RIS1, RISP, UQCR5; ubiquinol-cytochrom... 59.7 3e-09
cel:F42G8.12 isp-1; Iron-Sulfur Protein family member (isp-1);... 51.2 9e-07
xla:398972 arhgap21, arhgap21-B, rGAP, xrgap; Rho GTPase activ... 30.4 1.5
hsa:57214 CCSP1, TMEM2L; KIAA1199 30.0 2.1
mmu:80982 9930013L23Rik, 12H19.01.T7, 6330404C01Rik, AY007814;... 30.0 2.2
hsa:91368 CDKN2AIPNL, MGC13017; CDKN2A interacting protein N-t... 28.9 4.6
> tgo:TGME49_120220 ubiquinol-cytochrome c reductase domain-containing
protein (EC:1.10.2.2); K00411 ubiquinol-cytochrome
c reductase iron-sulfur subunit [EC:1.10.2.2]
Length=487
Score = 228 bits (581), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 104/137 (75%), Positives = 119/137 (86%), Gaps = 0/137 (0%)
Query 1 TSVWANPDEPAVVSVGRFAPDNFRAAGYAENAPNPTSINSDAHPDFRDYRLGSGSPDRRP 60
TSVW +P+EPA+VSV RFAPDNFRA G+AEN PNP S NSD+HPDFR+YRLG GS DRRP
Sbjct 265 TSVWHDPNEPAIVSVSRFAPDNFRAVGFAENVPNPESTNSDSHPDFREYRLGPGSVDRRP 324
Query 61 FMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTEIDLRPIEVGTTAVFKWRGK 120
F+YF+SASYFFI ASMMRS LCK VH+WWVS+D++AAGTTE+DLRPI+ G TAVFKWRGK
Sbjct 325 FVYFMSASYFFITASMMRSFLCKWVHYWWVSRDMLAAGTTEVDLRPIQEGMTAVFKWRGK 384
Query 121 PVFVRHRTPEEIEPCQG 137
PVFVRHRT E+I Q
Sbjct 385 PVFVRHRTAEDIAKAQA 401
> pfa:PF14_0373 ubiquinol-cytochrome c reductase iron-sulfur subunit,
putative; K00411 ubiquinol-cytochrome c reductase iron-sulfur
subunit [EC:1.10.2.2]
Length=355
Score = 206 bits (525), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 92/133 (69%), Positives = 110/133 (82%), Gaps = 0/133 (0%)
Query 1 TSVWANPDEPAVVSVGRFAPDNFRAAGYAENAPNPTSINSDAHPDFRDYRLGSGSPDRRP 60
T VW NP EPA+VS+G+F P NFR AGYAEN PNP SINSD HPDFR+YRL SG+ DRR
Sbjct 133 TDVWHNPKEPAIVSIGKFEPRNFRPAGYAENCPNPESINSDHHPDFREYRLRSGNEDRRS 192
Query 61 FMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTEIDLRPIEVGTTAVFKWRGK 120
FMYF+SASYFFI +S+MRS +CK VHF+W+SKDL+A GTTE+D+R + G V KWRGK
Sbjct 193 FMYFISASYFFIMSSIMRSAICKSVHFFWISKDLVAGGTTELDMRTVNPGEHVVIKWRGK 252
Query 121 PVFVRHRTPEEIE 133
PVFV+HRTPE+I+
Sbjct 253 PVFVKHRTPEDIQ 265
> bbo:BBOV_III009930 17.m07861; ubiquinol cytochrome c oxidoreductase
(EC:1.10.2.2); K00411 ubiquinol-cytochrome c reductase
iron-sulfur subunit [EC:1.10.2.2]
Length=332
Score = 138 bits (347), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 66/133 (49%), Positives = 92/133 (69%), Gaps = 1/133 (0%)
Query 1 TSVWANPDEPAVVSVGRFAPDNFRAAGYAENAPNPTSINSDAHPDFRDYRLGSGSPDRRP 60
TS W P EPA+VSVG+ PDN RA GYAENA P SI DA PDF +YRL G+ DRR
Sbjct 111 TSGWHLPGEPALVSVGKLGPDNQRAIGYAENAVVPDSIVPDAFPDFGEYRLPKGT-DRRA 169
Query 61 FMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTEIDLRPIEVGTTAVFKWRGK 120
+Y +SA+ F ++ RSL+CK++H++W+++D+ A+G+ E+++ + G KWR K
Sbjct 170 AIYIMSATAMFFFLALGRSLVCKIIHYFWIARDIAASGSVEVNVSQMIPGDQVTVKWRSK 229
Query 121 PVFVRHRTPEEIE 133
PVF++ RT EEIE
Sbjct 230 PVFIKRRTQEEIE 242
> tpv:TP04_0913 ubiquinol-cytochrome C reductase, iron-sulfur
subunit (EC:1.10.2.2); K00411 ubiquinol-cytochrome c reductase
iron-sulfur subunit [EC:1.10.2.2]
Length=339
Score = 127 bits (318), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 62/132 (46%), Positives = 94/132 (71%), Gaps = 1/132 (0%)
Query 1 TSVWANPDEPAVVSVGRFAPDNFRAAGYAENAPNPTSINSDAHPDFRDYRLGSGSPDRRP 60
TS W +P+EP +VSVG+ +P N R GYAEN P S ++ PDFR+YRL G+ DR+
Sbjct 118 TSGWYSPNEPTIVSVGKLSPANQRPIGYAENCVIPESTYPESFPDFREYRLPRGT-DRKA 176
Query 61 FMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTEIDLRPIEVGTTAVFKWRGK 120
+Y ++A++FF + +RSL+CK+VH++W+++D++AAG+ E+++ + G KWR K
Sbjct 177 TIYLMTATFFFFFFAFLRSLICKVVHYFWLTRDVVAAGSVEVNVGQMLPGDQITVKWRNK 236
Query 121 PVFVRHRTPEEI 132
PVF+R RTPEEI
Sbjct 237 PVFIRRRTPEEI 248
> ath:AT5G13430 ubiquinol-cytochrome C reductase iron-sulfur subunit,
mitochondrial, putative / Rieske iron-sulfur protein,
putative; K00411 ubiquinol-cytochrome c reductase iron-sulfur
subunit [EC:1.10.2.2]
Length=272
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 56/92 (60%), Gaps = 0/92 (0%)
Query 42 AHPDFRDYRLGSGSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTE 101
+ D R G P +R F YF+ + F+ AS++R L+ K++ SKD++A + E
Sbjct 92 VYDDHNHERYPPGDPSKRAFAYFVLSGGRFVYASVLRLLVLKLIVSMSASKDVLALASLE 151
Query 102 IDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
+DL IE GTT KWRGKPVF+R RT ++I+
Sbjct 152 VDLGSIEPGTTVTVKWRGKPVFIRRRTEDDIK 183
> ath:AT5G13440 ubiquinol-cytochrome C reductase iron-sulfur subunit,
mitochondrial, putative / Rieske iron-sulfur protein,
putative; K00411 ubiquinol-cytochrome c reductase iron-sulfur
subunit [EC:1.10.2.2]
Length=274
Score = 79.0 bits (193), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 56/92 (60%), Gaps = 0/92 (0%)
Query 42 AHPDFRDYRLGSGSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTE 101
+ D R G P +R F YF+ + F+ AS++R L+ K++ SKD++A + E
Sbjct 94 VYDDHNHERYPPGDPSKRAFAYFVLSGGRFVYASVLRLLVLKLIVSMSASKDVLALASLE 153
Query 102 IDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
+DL IE GTT KWRGKPVF+R RT ++I+
Sbjct 154 VDLGSIEPGTTVTVKWRGKPVFIRRRTEDDIK 185
> dre:406359 uqcrfs1, MGC174806, fb78f10, wu:fb05d11, wu:fb78f10,
wu:fj05e12, zgc:73109, zgc:85614; ubiquinol-cytochrome c
reductase, Rieske iron-sulfur polypeptide 1 (EC:1.10.2.2);
K00411 ubiquinol-cytochrome c reductase iron-sulfur subunit
[EC:1.10.2.2]
Length=273
Score = 71.6 bits (174), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 40/104 (38%), Positives = 52/104 (50%), Gaps = 10/104 (9%)
Query 44 PDFRDYR---------LGSGSPD-RRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKD 93
PDF DYR S D RR F Y ++ S +G ++++ + V S D
Sbjct 85 PDFSDYRRPEVLNPNKQSQESGDARRAFSYLMTGSTLVVGVYTAKTVVTQFVSSMSASAD 144
Query 94 LMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIEPCQG 137
++A EI L I G FKWRGKP+FVRHRT +EIE G
Sbjct 145 VLALSKIEIKLADIPEGKNMTFKWRGKPLFVRHRTEKEIETEAG 188
> dre:100334857 ubiquinol-cytochrome c reductase, Rieske iron-sulfur
polypeptide 1-like; K00411 ubiquinol-cytochrome c reductase
iron-sulfur subunit [EC:1.10.2.2]
Length=273
Score = 71.6 bits (174), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 40/104 (38%), Positives = 52/104 (50%), Gaps = 10/104 (9%)
Query 44 PDFRDYR---------LGSGSPD-RRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKD 93
PDF DYR S D RR F Y ++ S +G ++++ + V S D
Sbjct 85 PDFSDYRRPEVLNPNKQSQESGDARRAFSYLMTGSTLVVGVYTAKTVVTQFVSSMSASAD 144
Query 94 LMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIEPCQG 137
++A EI L I G FKWRGKP+FVRHRT +EIE G
Sbjct 145 VLALSKIEIKLADIPEGKNMTFKWRGKPLFVRHRTEKEIETEAG 188
> sce:YEL024W RIP1; Rip1p (EC:1.10.2.2); K00411 ubiquinol-cytochrome
c reductase iron-sulfur subunit [EC:1.10.2.2]
Length=215
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 50/91 (54%), Gaps = 1/91 (1%)
Query 44 PDFRDYRLGSGSPDR-RPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKDLMAAGTTEI 102
P+F D + D+ R + YF+ + + ++ +S + + + D++A E+
Sbjct 37 PNFDDVLKENNDADKGRSYAYFMVGAMGLLSSAGAKSTVETFISSMTATADVLAMAKVEV 96
Query 103 DLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
+L I +G V KW+GKPVF+RHRTP EI+
Sbjct 97 NLAAIPLGKNVVVKWQGKPVFIRHRTPHEIQ 127
> xla:779413 hypothetical protein MGC154858; K00411 ubiquinol-cytochrome
c reductase iron-sulfur subunit [EC:1.10.2.2]
Length=273
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 49/100 (49%), Gaps = 10/100 (10%)
Query 44 PDFRDYRL----------GSGSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKD 93
PDF DYR + S R+ F Y ++ + A ++ + + V S D
Sbjct 85 PDFSDYRRPEVLDSTKSSQTSSDSRKTFSYLVTGATAVASAYAAKNAVSQFVTSMSASAD 144
Query 94 LMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
++A EI L I G FKWRGKP+FVRHRT +EI+
Sbjct 145 VLAMSKIEIKLSDIPEGKNMAFKWRGKPLFVRHRTAKEID 184
> mmu:66694 Uqcrfs1, 4430402G14Rik, AI875505; ubiquinol-cytochrome
c reductase, Rieske iron-sulfur polypeptide 1 (EC:1.10.2.2);
K00411 ubiquinol-cytochrome c reductase iron-sulfur subunit
[EC:1.10.2.2]
Length=274
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 36/100 (36%), Positives = 50/100 (50%), Gaps = 10/100 (10%)
Query 44 PDFRDYRLGS----------GSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKD 93
PDF DYR S R+ F Y ++A+ A ++++ + V S D
Sbjct 86 PDFSDYRRAEVLDSTKSSKESSEARKGFSYLVTATTTVGVAYAAKNVVSQFVSSMSASAD 145
Query 94 LMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
++A EI L I G FKWRGKP+FVRHRT +EI+
Sbjct 146 VLAMSKIEIKLSDIPEGKNMAFKWRGKPLFVRHRTKKEID 185
> xla:379942 uqcrfs1, MGC53134; ubiquinol-cytochrome c reductase,
Rieske iron-sulfur polypeptide 1 (EC:1.10.2.2); K00411 ubiquinol-cytochrome
c reductase iron-sulfur subunit [EC:1.10.2.2]
Length=268
Score = 60.8 bits (146), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 49/100 (49%), Gaps = 10/100 (10%)
Query 44 PDFRDYRL----------GSGSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKD 93
PDF DYR + S R+ F Y ++ + A ++ + + V S D
Sbjct 80 PDFSDYRRPEVLDSTKSSQTSSDSRKSFSYLVTGATAVATAYAAKNAVSQFVTSMSASAD 139
Query 94 LMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
++A EI L I G FKWRGKP+FVRHRT +EI+
Sbjct 140 VLAMSKIEIKLSDIPEGKNMAFKWRGKPLFVRHRTAKEID 179
> hsa:7386 UQCRFS1, RIP1, RIS1, RISP, UQCR5; ubiquinol-cytochrome
c reductase, Rieske iron-sulfur polypeptide 1 (EC:1.10.2.2);
K00411 ubiquinol-cytochrome c reductase iron-sulfur subunit
[EC:1.10.2.2]
Length=274
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 49/100 (49%), Gaps = 10/100 (10%)
Query 44 PDFRDYR----LGS------GSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHFWWVSKD 93
PDF +YR L S S R+ F Y ++ A ++ + + V S D
Sbjct 86 PDFSEYRRLEVLDSTKSSRESSEARKGFSYLVTGVTTVGVAYAAKNAVTQFVSSMSASAD 145
Query 94 LMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEIE 133
++A EI L I G FKWRGKP+FVRHRT +EIE
Sbjct 146 VLALAKIEIKLSDIPEGKNMAFKWRGKPLFVRHRTQKEIE 185
> cel:F42G8.12 isp-1; Iron-Sulfur Protein family member (isp-1);
K00411 ubiquinol-cytochrome c reductase iron-sulfur subunit
[EC:1.10.2.2]
Length=276
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 11/100 (11%)
Query 44 PDFRDYRLGSGSPDRRPFM-------YFLSASYFFIGASMM----RSLLCKMVHFWWVSK 92
PD +YR S + P +A Y+ G + + ++ +V + ++
Sbjct 86 PDMSNYRRDSTKNTQVPARDTEDQRRALPTALYYGAGGVLSLWAGKEVVQTLVSYKAMAA 145
Query 93 DLMAAGTTEIDLRPIEVGTTAVFKWRGKPVFVRHRTPEEI 132
D A + EI++ I G T F+WRGKPVFV+HRT EI
Sbjct 146 DQRALASIEINMADIPEGKTKTFEWRGKPVFVKHRTKAEI 185
> xla:398972 arhgap21, arhgap21-B, rGAP, xrgap; Rho GTPase activating
protein 21
Length=1902
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 23/57 (40%), Positives = 29/57 (50%), Gaps = 7/57 (12%)
Query 14 SVGRFAPDNF--RAAGYAENAPNPT----SINSDAHPDFRDYRLGSGSPD-RRPFMY 63
S G A + F R A +AE +P+PT S S H D+RDY+ D RR MY
Sbjct 335 SYGGLAENMFSTRPAAHAEESPSPTNHYASPGSHQHIDWRDYKTYKEYIDNRRMLMY 391
> hsa:57214 CCSP1, TMEM2L; KIAA1199
Length=1361
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 33/68 (48%), Gaps = 14/68 (20%)
Query 10 PAVVSVGRFAPDNFRAAGYAENAPNPTSINSDAHPDFRDYRL--------GSGSPDRRPF 61
P SVG ++P GY+E+ P N+ AH ++R + + + D+RPF
Sbjct 703 PTGPSVGMYSP------GYSEHIPLGKFYNNRAHSNYRAGMIIDNGVKTTEASAKDKRPF 756
Query 62 MYFLSASY 69
+ +SA Y
Sbjct 757 LSIISARY 764
> mmu:80982 9930013L23Rik, 12H19.01.T7, 6330404C01Rik, AY007814;
RIKEN cDNA 9930013L23 gene
Length=1361
Score = 30.0 bits (66), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 33/68 (48%), Gaps = 14/68 (20%)
Query 10 PAVVSVGRFAPDNFRAAGYAENAPNPTSINSDAHPDFRDYRL--------GSGSPDRRPF 61
P SVG ++P GY+E+ P N+ AH ++R + + + D+RPF
Sbjct 703 PTGPSVGMYSP------GYSEHIPLGKFYNNRAHSNYRAGMIIDNGVKTTEASAKDKRPF 756
Query 62 MYFLSASY 69
+ +SA Y
Sbjct 757 LSIISARY 764
> hsa:91368 CDKN2AIPNL, MGC13017; CDKN2A interacting protein N-terminal
like
Length=116
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 44 PDFRDYRLGSGSPDRRPFMYFLSASYFFIGASMMRSLLCKMVHF 87
PD+RD GSG D+ + + A++ F+G S + LL K++
Sbjct 49 PDYRDPPDGSGRLDQLLSLSMVWANHLFLGCSYNKDLLDKVMEM 92
Lambda K H
0.322 0.137 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2421919804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40