bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4995_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
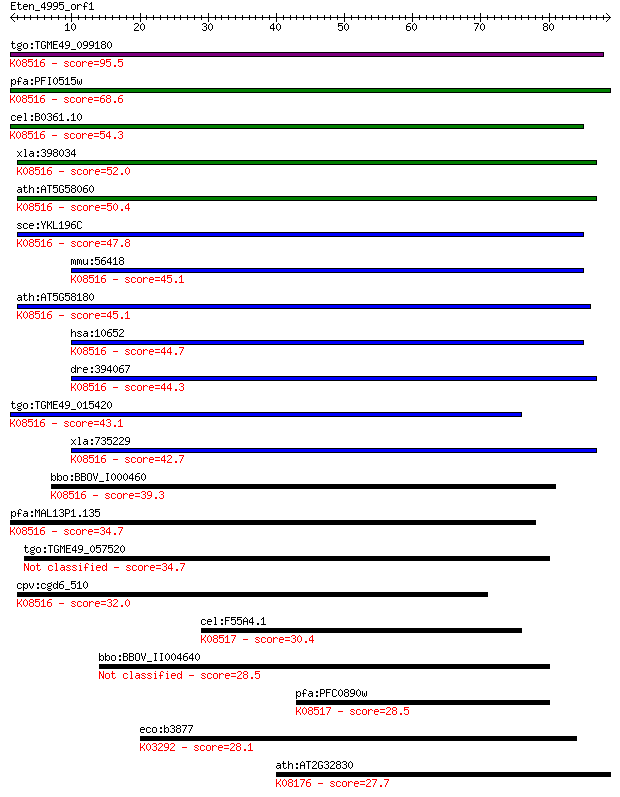
tgo:TGME49_099180 hypothetical protein ; K08516 synaptobrevin ... 95.5 3e-20
pfa:PFI0515w PfYkt6.1; SNARE protein, putative; K08516 synapto... 68.6 4e-12
cel:B0361.10 hypothetical protein; K08516 synaptobrevin homolo... 54.3 8e-08
xla:398034 ykt6-a, ykt6, ykt6a; YKT6 v-SNARE homolog (EC:2.3.1... 52.0 4e-07
ath:AT5G58060 YKT61; YKT61; K08516 synaptobrevin homolog YKT6 50.4 1e-06
sce:YKL196C YKT6; Vesicle membrane protein (v-SNARE) with acyl... 47.8 8e-06
mmu:56418 Ykt6, 0610042I15Rik, 1810013M05Rik, AW105923; YKT6 h... 45.1 5e-05
ath:AT5G58180 ATYKT62; K08516 synaptobrevin homolog YKT6 45.1 5e-05
hsa:10652 YKT6; YKT6 v-SNARE homolog (S. cerevisiae); K08516 s... 44.7 8e-05
dre:394067 MGC55536, Ykt6; zgc:55536 (EC:2.3.1.-); K08516 syna... 44.3 1e-04
tgo:TGME49_015420 SNARE protein, putative ; K08516 synaptobrev... 43.1 2e-04
xla:735229 ykt6-b, ykt6b; YKT6 v-SNARE homolog (EC:2.3.1.-); K... 42.7 3e-04
bbo:BBOV_I000460 16.m00764; hypothetical protein; K08516 synap... 39.3 0.003
pfa:MAL13P1.135 PfYkt6.2; SNARE protein, putative; K08516 syna... 34.7 0.068
tgo:TGME49_057520 synaptobrevin-like protein, putative 34.7 0.079
cpv:cgd6_510 synaptobrevin/VAMP-like protein ; K08516 synaptob... 32.0 0.50
cel:F55A4.1 hypothetical protein; K08517 vesicle transport pro... 30.4 1.3
bbo:BBOV_II004640 18.m06388; vesicle transport protein 28.5 5.9
pfa:PFC0890w PfSec22; SNARE protein; K08517 vesicle transport ... 28.5 6.1
eco:b3877 yihP, ECK3870, JW3848; predicted transporter; K03292... 28.1 6.3
ath:AT2G32830 PHT5; PHT5; inorganic phosphate transmembrane tr... 27.7 9.5
> tgo:TGME49_099180 hypothetical protein ; K08516 synaptobrevin
homolog YKT6
Length=211
Score = 95.5 bits (236), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 39/87 (44%), Positives = 66/87 (75%), Gaps = 0/87 (0%)
Query 1 AFSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYP 60
++FY R+++KE KF+ +TAIPR+ V +++ +EH+G AF++R+ +SLA +A+GD YP
Sbjct 29 GYAFYQRHSMKEACKFLARTAIPRVGVETREVIEHDGSVAFLYRFFDSLAVVAIGDANYP 88
Query 61 RRVAFACLNEVYLQFTQSVPASKWKSL 87
RVAF L+E++ +FT++VP + W S+
Sbjct 89 SRVAFRLLHEIHEKFTRAVPVALWSSV 115
> pfa:PFI0515w PfYkt6.1; SNARE protein, putative; K08516 synaptobrevin
homolog YKT6
Length=199
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 50/88 (56%), Gaps = 0/88 (0%)
Query 1 AFSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYP 60
++SF + KE FV +T R+ N+++ + HE F F+Y +++ + + +YP
Sbjct 26 SYSFIKKKAFKEAAFFVARTIPSRIEYNTKEIITHENNTVFAFKYEDNICPIVIATDDYP 85
Query 61 RRVAFACLNEVYLQFTQSVPASKWKSLE 88
RVAF +NE+Y F ++P +W S++
Sbjct 86 ERVAFYMINEIYRDFISTIPKEEWSSVK 113
> cel:B0361.10 hypothetical protein; K08516 synaptobrevin homolog
YKT6
Length=201
Score = 54.3 bits (129), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 45/84 (53%), Gaps = 0/84 (0%)
Query 1 AFSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYP 60
+FSF+ R +V+E + F K + R + ++ V+ + + L+A+ V D EY
Sbjct 29 SFSFFQRGSVQEFMTFTAKLLVERSGLGARSSVKENEYLVHCYVRNDGLSAVCVTDAEYQ 88
Query 61 RRVAFACLNEVYLQFTQSVPASKW 84
+RVA + L V FT VPA++W
Sbjct 89 QRVAMSFLGRVLDDFTTRVPATQW 112
> xla:398034 ykt6-a, ykt6, ykt6a; YKT6 v-SNARE homolog (EC:2.3.1.-);
K08516 synaptobrevin homolog YKT6
Length=198
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 46/86 (53%), Gaps = 2/86 (2%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEV-EHEGLCAFVFRYPNSLAALAVGDKEYP 60
FSF+ R++++E + F + + R S+ V E E LC V+ +SLA + + D EYP
Sbjct 28 FSFFQRSSIQEFMAFTSQLIVERSDKGSRSSVKEQEYLC-HVYVRNDSLAGVVIADNEYP 86
Query 61 RRVAFACLNEVYLQFTQSVPASKWKS 86
RV F L +V +F+ V W S
Sbjct 87 PRVCFTLLEKVLEEFSTQVDRIDWPS 112
> ath:AT5G58060 YKT61; YKT61; K08516 synaptobrevin homolog YKT6
Length=221
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 50/106 (47%), Gaps = 22/106 (20%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAF-VFRYP-------------- 46
F ++ R++VKE + FV +T R P + +Q V+HEG F + P
Sbjct 30 FGYFQRSSVKEFVVFVGRTVASRTPPSQRQSVQHEGCAPFLILDLPGLCPGFNFLRFYLI 89
Query 47 ------NSLAALAVGDKEYPRRVAFACLNEVYLQFTQSVPASKWKS 86
N L A+ D YP R AF+ LN+V ++ +S S W+S
Sbjct 90 VHAYNRNGLCAVGFMDDHYPVRSAFSLLNQVLDEYQKSFGES-WRS 134
> sce:YKL196C YKT6; Vesicle membrane protein (v-SNARE) with acyltransferase
activity; involved in trafficking to and within
the Golgi, endocytic trafficking to the vacuole, and vacuolar
fusion; membrane localization due to prenylation at the
carboxy-terminus (EC:2.3.1.-); K08516 synaptobrevin homolog
YKT6
Length=200
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 21/83 (25%), Positives = 40/83 (48%), Gaps = 0/83 (0%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPR 61
F F+ R++V + + F +T R +Q +E V+ + + + DKEYP
Sbjct 28 FGFFERSSVGQFMTFFAETVASRTGAGQRQSIEEGNYIGHVYARSEGICGVLITDKEYPV 87
Query 62 RVAFACLNEVYLQFTQSVPASKW 84
R A+ LN++ ++ + P +W
Sbjct 88 RPAYTLLNKILDEYLVAHPKEEW 110
> mmu:56418 Ykt6, 0610042I15Rik, 1810013M05Rik, AW105923; YKT6
homolog (S. Cerevisiae); K08516 synaptobrevin homolog YKT6
Length=198
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 27/76 (35%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Query 10 VKEGLKFVVKTAIPRLPVNSQQEV-EHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFACL 68
V+E + F + + R S+ V E E LC V+ +SLA + + D EYP RVAF L
Sbjct 36 VQEFMTFTSQLIVERSGKGSRASVKEQEYLC-HVYVRSDSLAGVVIADSEYPSRVAFTLL 94
Query 69 NEVYLQFTQSVPASKW 84
+V +F++ V W
Sbjct 95 EKVLDEFSKQVDRIDW 110
> ath:AT5G58180 ATYKT62; K08516 synaptobrevin homolog YKT6
Length=199
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 41/84 (48%), Gaps = 3/84 (3%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPR 61
FSFY R+ +E + F+ +T R P +Q V+HE + N L A+ D YP
Sbjct 33 FSFY-RSNFEEFIVFIARTVARRTPPGQRQSVKHEEYKVHAYNI-NGLCAVGFMDDHYPV 90
Query 62 RVAFACLNEVYLQFTQSVPASKWK 85
R AF+ LN+V L Q W+
Sbjct 91 RSAFSLLNQV-LDVYQKDYGDTWR 113
> hsa:10652 YKT6; YKT6 v-SNARE homolog (S. cerevisiae); K08516
synaptobrevin homolog YKT6
Length=198
Score = 44.7 bits (104), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 10 VKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFACLN 69
V+E + F + + R ++ V+ + V+ +SLA + + D EYP RVAF L
Sbjct 36 VQEFMTFTSQLIVERSSKGTRASVKEQDYLCHVYVRNDSLAGVVIADNEYPSRVAFTLLE 95
Query 70 EVYLQFTQSVPASKW 84
+V +F++ V W
Sbjct 96 KVLDEFSKQVDRIDW 110
> dre:394067 MGC55536, Ykt6; zgc:55536 (EC:2.3.1.-); K08516 synaptobrevin
homolog YKT6
Length=198
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 39/78 (50%), Gaps = 2/78 (2%)
Query 10 VKEGLKFVVKTAIPRLPVNSQQEV-EHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFACL 68
V+E + F + R + S+ V E E LC R N L + + D EYP RV F L
Sbjct 36 VQEFMTFTSALIVERSALGSRASVKEQEYLCHVYVRNDN-LGGVVIADSEYPSRVCFTLL 94
Query 69 NEVYLQFTQSVPASKWKS 86
++V +F++ V + W S
Sbjct 95 DKVLDEFSRQVNSIDWPS 112
> tgo:TGME49_015420 SNARE protein, putative ; K08516 synaptobrevin
homolog YKT6
Length=263
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 40/78 (51%), Gaps = 4/78 (5%)
Query 1 AFSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHE---GLCAFVFRYPNSLAALAVGDK 57
+F F+ R+T+KE + F + R P+ +Q VE E G C VF + + LAA +
Sbjct 54 SFPFFHRSTMKEHIVFHSRLICARTPLGRRQVVEFEQNIGHC-HVFVHSSGLAATVLSTA 112
Query 58 EYPRRVAFACLNEVYLQF 75
YP RVAF + + F
Sbjct 113 AYPMRVAFGLITQALRGF 130
> xla:735229 ykt6-b, ykt6b; YKT6 v-SNARE homolog (EC:2.3.1.-);
K08516 synaptobrevin homolog YKT6
Length=198
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 39/78 (50%), Gaps = 2/78 (2%)
Query 10 VKEGLKFVVKTAIPRLPVNSQQEV-EHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFACL 68
V+E + F + + R S+ V E E LC V+ +SLA + + D EYP RV F L
Sbjct 36 VQEFMTFTSQLIVERSDKGSRSSVKEQEYLC-HVYVRNDSLAGVVIADNEYPPRVCFTLL 94
Query 69 NEVYLQFTQSVPASKWKS 86
+V +F+ V W S
Sbjct 95 EKVLEEFSTQVDRIDWPS 112
> bbo:BBOV_I000460 16.m00764; hypothetical protein; K08516 synaptobrevin
homolog YKT6
Length=202
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/74 (29%), Positives = 31/74 (41%), Gaps = 0/74 (0%)
Query 7 RNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFA 66
R TVK F + RL V E F +R+ + A+ + K YP RVAF+
Sbjct 35 RGTVKSVAVFTAREIAGRLNQGENVAVHEENFDVFAYRWDVGVCAICICGKGYPERVAFS 94
Query 67 CLNEVYLQFTQSVP 80
L + +F P
Sbjct 95 LLQLAFFEFITKYP 108
> pfa:MAL13P1.135 PfYkt6.2; SNARE protein, putative; K08516 synaptobrevin
homolog YKT6
Length=221
Score = 34.7 bits (78), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 19/80 (23%), Positives = 40/80 (50%), Gaps = 3/80 (3%)
Query 1 AFSFYXRNTVKEGLKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPN---SLAALAVGDK 57
+F F+ R+++KE + F + R +++ +E E + Y N +++ L +
Sbjct 44 SFPFFHRSSLKEHIYFHARLVCGRTQKGTREVIELESGIGHLHIYTNKENNISVLVLSTS 103
Query 58 EYPRRVAFACLNEVYLQFTQ 77
YP R+AF+ ++ + F Q
Sbjct 104 SYPLRIAFSLIDLTHKLFAQ 123
> tgo:TGME49_057520 synaptobrevin-like protein, putative
Length=266
Score = 34.7 bits (78), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 40/85 (47%), Gaps = 8/85 (9%)
Query 3 SFYXRNTVKEGLKF------VVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNS--LAALAV 54
+FY R T KE V+KTA +P ++ +G F+ P L L
Sbjct 54 TFYNRLTSKEKNALAPLFPQVLKTAPDAMPGLRRKFPVDDGGIMFLASDPTGSFLFGLYT 113
Query 55 GDKEYPRRVAFACLNEVYLQFTQSV 79
DK+YP RVAF L EV+ +++V
Sbjct 114 HDKQYPERVAFTFLTEVHQLVSEAV 138
> cpv:cgd6_510 synaptobrevin/VAMP-like protein ; K08516 synaptobrevin
homolog YKT6
Length=225
Score = 32.0 bits (71), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 18/75 (24%), Positives = 35/75 (46%), Gaps = 10/75 (13%)
Query 2 FSFYXRNTVKEGLKFVVKTAIPRLPVNSQ------QEVEHEGLCAFVFRYPNSLAALAVG 55
++F+ R T++E + F + R+ ++ Q++ H ++ N L +
Sbjct 28 YNFFQRKTIQEHIAFHSRLLCSRVQQGNRVTVTFPQDIGH----CHIYISNNGLGICCIT 83
Query 56 DKEYPRRVAFACLNE 70
+YP R AF+ LNE
Sbjct 84 SPDYPVRCAFSLLNE 98
> cel:F55A4.1 hypothetical protein; K08517 vesicle transport protein
SEC22
Length=214
Score = 30.4 bits (67), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 29/49 (59%), Gaps = 5/49 (10%)
Query 29 SQQEVEHEGLCAFVFRY--PNSLAALAVGDKEYPRRVAFACLNEVYLQF 75
+QQ VE FVF Y ++ AL + D+ +PR+VAF L+++ +F
Sbjct 50 AQQSVES---GPFVFHYIIVQNICALVLCDRNFPRKVAFQYLSDIGQEF 95
> bbo:BBOV_II004640 18.m06388; vesicle transport protein
Length=194
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 0/66 (0%)
Query 14 LKFVVKTAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFACLNEVYL 73
+K +T L NS + EG ++ + + +A L V YP+++AF L EV
Sbjct 43 IKMYARTICRSLQPNSLKSSIKEGEYSYHYTVEDGVAYLTVVPSTYPKKLAFLYLAEVCT 102
Query 74 QFTQSV 79
F++ +
Sbjct 103 AFSKEL 108
> pfa:PFC0890w PfSec22; SNARE protein; K08517 vesicle transport
protein SEC22
Length=221
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 43 FRYPNSLAALAVGDKEYPRRVAFACLNEVYLQFTQSV 79
F N +A +AV YP+++AF LN++ QF + +
Sbjct 57 FLIENGIAYIAVFPVTYPKKLAFLFLNDICKQFNEEL 93
> eco:b3877 yihP, ECK3870, JW3848; predicted transporter; K03292
glycoside/pentoside/hexuronide:cation symporter, GPH family
Length=461
Score = 28.1 bits (61), Expect = 6.3, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 34/68 (50%), Gaps = 12/68 (17%)
Query 20 TAIPRLPVNSQQEVEHEGLCAFVFRYPNSLAALAVGDKEYPRRVAFAC---LNE-VYLQF 75
T I +P +Q + EGL +F YP++LA + + VA C LNE +Y++
Sbjct 399 TQIGYVPNVAQADHTIEGLRQLIFIYPSALAVVTI--------VAMGCFYSLNEKMYVRI 450
Query 76 TQSVPASK 83
+ + A K
Sbjct 451 VEEIEARK 458
> ath:AT2G32830 PHT5; PHT5; inorganic phosphate transmembrane
transporter/ phosphate transmembrane transporter; K08176 MFS
transporter, PHS family, inorganic phosphate transporter
Length=542
Score = 27.7 bits (60), Expect = 9.5, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 25/56 (44%), Gaps = 7/56 (12%)
Query 40 AFVFRYPNSLAALAVGDKEYPRRVA-------FACLNEVYLQFTQSVPASKWKSLE 88
AF F Y + D YP + AC+N + + FT VP SK KSLE
Sbjct 458 AFGFLYAAQSSDSEKTDAGYPPGIGVRNSLLMLACVNFLGIVFTLLVPESKGKSLE 513
Lambda K H
0.322 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2021645584
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40