bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4929_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
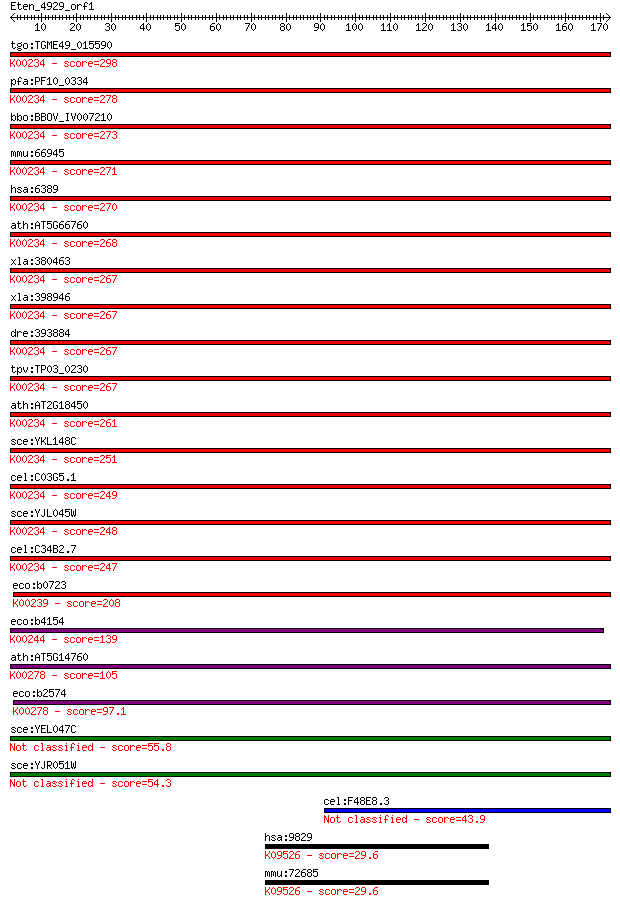
tgo:TGME49_015590 succinate dehydrogenase (ubiquinone) flavopr... 298 7e-81
pfa:PF10_0334 flavoprotein subunit of succinate dehydrogenase ... 278 7e-75
bbo:BBOV_IV007210 23.m06369; succinate dehydrogenase alpha sub... 273 2e-73
mmu:66945 Sdha, 1500032O14Rik, 2310034D06Rik, 4921513A11, C810... 271 1e-72
hsa:6389 SDHA, CMD1GG, FP, SDH1, SDH2, SDHF; succinate dehydro... 270 1e-72
ath:AT5G66760 SDH1-1; SDH1-1; ATP binding / succinate dehydrog... 268 5e-72
xla:380463 sdha, MGC53323, sdha-b; succinate dehydrogenase com... 267 1e-71
xla:398946 MGC68518, sdha-a; succinate dehydrogenase [ubiquino... 267 1e-71
dre:393884 sdha, MGC56051, im:7141001, zgc:56051; succinate de... 267 1e-71
tpv:TP03_0230 succinate dehydrogenase flavoprotein subunit (EC... 267 1e-71
ath:AT2G18450 SDH1-2; SDH1-2; succinate dehydrogenase (EC:1.3.... 261 9e-70
sce:YKL148C SDH1; Flavoprotein subunit of succinate dehydrogen... 251 7e-67
cel:C03G5.1 sdha-1; Succinate DeHydrogenase complex subunit A ... 249 4e-66
sce:YJL045W Minor succinate dehydrogenase isozyme; homologous ... 248 7e-66
cel:C34B2.7 sdha-2; Succinate DeHydrogenase complex subunit A ... 247 2e-65
eco:b0723 sdhA, ECK0712, JW0713; succinate dehydrogenase, flav... 208 8e-54
eco:b4154 frdA, ECK4150, JW4115; fumarate reductase (anaerobic... 139 4e-33
ath:AT5G14760 AO; AO (L-ASPARTATE OXIDASE); L-aspartate oxidas... 105 7e-23
eco:b2574 nadB, ECK2572, JW2558, nic, nicB; quinolinate syntha... 97.1 3e-20
sce:YEL047C Soluble fumarate reductase, required with isoenzym... 55.8 7e-08
sce:YJR051W OSM1; Fumarate reductase, catalyzes the reduction ... 54.3 2e-07
cel:F48E8.3 hypothetical protein 43.9 3e-04
hsa:9829 DNAJC6, DJC6, KIAA0473, MGC129914, MGC129915, MGC4843... 29.6 5.0
mmu:72685 Dnajc6, 2810027M23Rik, mKIAA0473; DnaJ (Hsp40) homol... 29.6 5.7
> tgo:TGME49_015590 succinate dehydrogenase (ubiquinone) flavoprotein
subunit, mitochondrial, putative (EC:1.3.5.1); K00234
succinate dehydrogenase (ubiquinone) flavoprotein subunit
[EC:1.3.5.1]
Length=669
Score = 298 bits (762), Expect = 7e-81, Method: Compositional matrix adjust.
Identities = 142/172 (82%), Positives = 155/172 (90%), Gaps = 1/172 (0%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGIFPAGCLITEGCRGEGGILRN +GEAFMARYAPTAKDLASRDVVSRSMTIEIR
Sbjct 309 VQFHPTGIFPAGCLITEGCRGEGGILRNGQGEAFMARYAPTAKDLASRDVVSRSMTIEIR 368
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCGP +DH HLDLTHL P TLH RLPGITETAKIFAGV+ K+PIPVLPTVHYNMGG
Sbjct 369 EGRGCGPNRDHMHLDLTHLPPATLHERLPGITETAKIFAGVNAEKEPIPVLPTVHYNMGG 428
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTNWR++V+ + K ++ +++GLYAAGEAACASVHGANRLGANSLLD
Sbjct 429 IPTNWRAQVLTTSRSKAGGPDK-IVQGLYAAGEAACASVHGANRLGANSLLD 479
> pfa:PF10_0334 flavoprotein subunit of succinate dehydrogenase
(EC:1.3.5.1); K00234 succinate dehydrogenase (ubiquinone)
flavoprotein subunit [EC:1.3.5.1]
Length=631
Score = 278 bits (711), Expect = 7e-75, Method: Compositional matrix adjust.
Identities = 135/176 (76%), Positives = 149/176 (84%), Gaps = 4/176 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+PAGCLITEGCRGEGGILRN EGEAFM RYAP AKDLASRDVVSR+MTIEI
Sbjct 266 VQFHPTGIYPAGCLITEGCRGEGGILRNKEGEAFMMRYAPKAKDLASRDVVSRAMTIEIN 325
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
E RGCGP DH +LDLTHL ETL RLPGI ETAKIFAGVDVTKQ IPVLPTVHYNMGG
Sbjct 326 EQRGCGPNADHIYLDLTHLPYETLKERLPGIMETAKIFAGVDVTKQYIPVLPTVHYNMGG 385
Query 121 IPTNWRSEV----IHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN++++V ++ K + E+ +++GLYAAGEAA ASVHGANRLGANSLLD
Sbjct 386 IPTNYKTQVLTQNVNFNKQTNKSNEDIIVKGLYAAGEAASASVHGANRLGANSLLD 441
> bbo:BBOV_IV007210 23.m06369; succinate dehydrogenase alpha subunit
(EC:1.3.5.1); K00234 succinate dehydrogenase (ubiquinone)
flavoprotein subunit [EC:1.3.5.1]
Length=624
Score = 273 bits (698), Expect = 2e-73, Method: Compositional matrix adjust.
Identities = 133/172 (77%), Positives = 141/172 (81%), Gaps = 8/172 (4%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+PAGCLITEGCRGEGGILRN EGE FM+RYAP AKDLASRDVVSR+MT EI
Sbjct 268 VQFHPTGIYPAGCLITEGCRGEGGILRNVEGEPFMSRYAPVAKDLASRDVVSRAMTCEIL 327
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCGP KDH +LDLTHL E +LPGITETAKIFAGVD KQ IPVLPTVHYNMGG
Sbjct 328 EGRGCGPNKDHIYLDLTHLSDEVFREKLPGITETAKIFAGVDARKQYIPVLPTVHYNMGG 387
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTNWR+E I+ K + GLYAAGE ACASVHGANRLGANSLLD
Sbjct 388 IPTNWRAEAINRDNSK--------IPGLYAAGETACASVHGANRLGANSLLD 431
> mmu:66945 Sdha, 1500032O14Rik, 2310034D06Rik, 4921513A11, C81073,
FP, SDH2, SDHF; succinate dehydrogenase complex, subunit
A, flavoprotein (Fp) (EC:1.3.5.1); K00234 succinate dehydrogenase
(ubiquinone) flavoprotein subunit [EC:1.3.5.1]
Length=664
Score = 271 bits (692), Expect = 1e-72, Method: Compositional matrix adjust.
Identities = 132/172 (76%), Positives = 147/172 (85%), Gaps = 5/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEGCRGEGGIL NS+GE FM RYAP AKDLASRDVVSRSMT+EIR
Sbjct 293 VQFHPTGIYGAGCLITEGCRGEGGILINSQGERFMERYAPVAKDLASRDVVSRSMTLEIR 352
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCGP KDH +L L HL PE L +RLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 353 EGRGCGPEKDHVYLQLHHLPPEQLATRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGG 412
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN++ +V+ V G+ + ++ GLYA GEAACASVHGANRLGANSLLD
Sbjct 413 IPTNYKGQVLKHVNGQDQ-----IVPGLYACGEAACASVHGANRLGANSLLD 459
> hsa:6389 SDHA, CMD1GG, FP, SDH1, SDH2, SDHF; succinate dehydrogenase
complex, subunit A, flavoprotein (Fp) (EC:1.3.5.1);
K00234 succinate dehydrogenase (ubiquinone) flavoprotein subunit
[EC:1.3.5.1]
Length=664
Score = 270 bits (691), Expect = 1e-72, Method: Compositional matrix adjust.
Identities = 132/172 (76%), Positives = 147/172 (85%), Gaps = 5/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEGCRGEGGIL NS+GE FM RYAP AKDLASRDVVSRSMT+EIR
Sbjct 293 VQFHPTGIYGAGCLITEGCRGEGGILINSQGERFMERYAPVAKDLASRDVVSRSMTLEIR 352
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCGP KDH +L L HL PE L +RLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 353 EGRGCGPEKDHVYLQLHHLPPEQLATRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGG 412
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN++ +V+ V G+ + ++ GLYA GEAACASVHGANRLGANSLLD
Sbjct 413 IPTNYKGQVLRHVNGQDQ-----IVPGLYACGEAACASVHGANRLGANSLLD 459
> ath:AT5G66760 SDH1-1; SDH1-1; ATP binding / succinate dehydrogenase
(EC:1.3.5.1); K00234 succinate dehydrogenase (ubiquinone)
flavoprotein subunit [EC:1.3.5.1]
Length=634
Score = 268 bits (686), Expect = 5e-72, Method: Compositional matrix adjust.
Identities = 135/172 (78%), Positives = 145/172 (84%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEG RGEGGILRNSEGE FM RYAPTAKDLASRDVVSRSMT+EIR
Sbjct 282 VQFHPTGIYGAGCLITEGSRGEGGILRNSEGERFMERYAPTAKDLASRDVVSRSMTMEIR 341
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG GP KDH +L L HL PE L RLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 342 EGRGVGPHKDHIYLHLNHLPPEVLKERLPGISETAAIFAGVDVTKEPIPVLPTVHYNMGG 401
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN+ EV+ +KG + V+ GL AAGEAACASVHGANRLGANSLLD
Sbjct 402 IPTNYHGEVV-TIKGDD---PDAVIPGLMAAGEAACASVHGANRLGANSLLD 449
> xla:380463 sdha, MGC53323, sdha-b; succinate dehydrogenase complex,
subunit A, flavoprotein (Fp) (EC:1.3.5.1); K00234 succinate
dehydrogenase (ubiquinone) flavoprotein subunit [EC:1.3.5.1]
Length=665
Score = 267 bits (683), Expect = 1e-71, Method: Compositional matrix adjust.
Identities = 134/172 (77%), Positives = 144/172 (83%), Gaps = 5/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEGCRGEGGIL NSEGE FM RYAP AKDLASRDVVSRSMTIE+R
Sbjct 296 VQFHPTGIYGAGCLITEGCRGEGGILINSEGERFMERYAPVAKDLASRDVVSRSMTIEMR 355
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCG KDH +L L HL P L SRLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 356 EGRGCGKDKDHVYLQLHHLPPSQLASRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGG 415
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN++ +VI V G+ R V+ GLYA GEAA ASVHGANRLGANSLLD
Sbjct 416 IPTNYKGQVITHVNGEDR-----VVPGLYACGEAASASVHGANRLGANSLLD 462
> xla:398946 MGC68518, sdha-a; succinate dehydrogenase [ubiquinone]
flavoprotein subunit A, mitochondrial precursor (EC:1.3.5.1);
K00234 succinate dehydrogenase (ubiquinone) flavoprotein
subunit [EC:1.3.5.1]
Length=665
Score = 267 bits (683), Expect = 1e-71, Method: Compositional matrix adjust.
Identities = 134/172 (77%), Positives = 144/172 (83%), Gaps = 5/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEGCRGEGGIL NSEGE FM RYAP AKDLASRDVVSRSMTIEIR
Sbjct 296 VQFHPTGIYGAGCLITEGCRGEGGILINSEGERFMERYAPVAKDLASRDVVSRSMTIEIR 355
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCG KDH +L L HL P L SRLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 356 EGRGCGKDKDHVYLQLHHLPPSQLASRLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGG 415
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN++ +VI V G+ R V+ GLY+ GEAA ASVHGANRLGANSLLD
Sbjct 416 IPTNYKGQVITHVNGEDR-----VVPGLYSCGEAASASVHGANRLGANSLLD 462
> dre:393884 sdha, MGC56051, im:7141001, zgc:56051; succinate
dehydrogenase complex, subunit A, flavoprotein (Fp) (EC:1.3.5.1);
K00234 succinate dehydrogenase (ubiquinone) flavoprotein
subunit [EC:1.3.5.1]
Length=661
Score = 267 bits (682), Expect = 1e-71, Method: Compositional matrix adjust.
Identities = 133/172 (77%), Positives = 144/172 (83%), Gaps = 5/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEGCRGEGGIL NSEGE FM RYAP AKDLASRDVVSRSMTIEIR
Sbjct 290 VQFHPTGIYGAGCLITEGCRGEGGILINSEGERFMERYAPNAKDLASRDVVSRSMTIEIR 349
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG GP KDH HL L HL P+ L +RLPGI+ETA IFAGVDVTK+PIPVLPTVHYNMGG
Sbjct 350 EGRGVGPDKDHVHLQLHHLPPQQLAARLPGISETAMIFAGVDVTKEPIPVLPTVHYNMGG 409
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN++ +VI G ++ V+ GLYA GEA CASVHGANRLGANSLLD
Sbjct 410 IPTNYKGQVITYKDG-----QDHVVPGLYACGEAGCASVHGANRLGANSLLD 456
> tpv:TP03_0230 succinate dehydrogenase flavoprotein subunit (EC:1.3.5.1);
K00234 succinate dehydrogenase (ubiquinone) flavoprotein
subunit [EC:1.3.5.1]
Length=658
Score = 267 bits (682), Expect = 1e-71, Method: Compositional matrix adjust.
Identities = 133/200 (66%), Positives = 150/200 (75%), Gaps = 28/200 (14%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+PAGCLITEGCRGEGGILRNSEGEAFMARYAP AKDLASRDVVSRSMTIEI
Sbjct 263 VQFHPTGIYPAGCLITEGCRGEGGILRNSEGEAFMARYAPVAKDLASRDVVSRSMTIEIN 322
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRGCGP KDH +LDLTHL + +LPGITET+KIFAGVD +K+ IPVLPTVHYNMGG
Sbjct 323 EGRGCGPNKDHLYLDLTHLSEDVFREKLPGITETSKIFAGVDASKEYIPVLPTVHYNMGG 382
Query 121 IPTNWRSEVIHCV-KGKGRM---------------------------AEECVLEGLYAAG 152
+PTN+++EV+ G G + + V+ GLY+AG
Sbjct 383 VPTNYKTEVVTTSPTGSGSSGVTAGNFLGDKVKGGKKKKMTVPKPTNSGDTVVYGLYSAG 442
Query 153 EAACASVHGANRLGANSLLD 172
E+ACASVHGANRLGANSLLD
Sbjct 443 ESACASVHGANRLGANSLLD 462
> ath:AT2G18450 SDH1-2; SDH1-2; succinate dehydrogenase (EC:1.3.5.1);
K00234 succinate dehydrogenase (ubiquinone) flavoprotein
subunit [EC:1.3.5.1]
Length=632
Score = 261 bits (667), Expect = 9e-70, Method: Compositional matrix adjust.
Identities = 131/172 (76%), Positives = 145/172 (84%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEG RGEGGILRNSEGE FM RYAPTA+DLASRDVVSRSMT+EIR
Sbjct 280 VQFHPTGIYGAGCLITEGARGEGGILRNSEGEKFMDRYAPTARDLASRDVVSRSMTMEIR 339
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
+GRG GP KD+ +L L HL PE L RLPGI+ETA IFAGVDVT++PIPVLPTVHYNMGG
Sbjct 340 QGRGAGPMKDYLYLYLNHLPPEVLKERLPGISETAAIFAGVDVTREPIPVLPTVHYNMGG 399
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPTN+ EVI ++G + V+ GL AAGEAACASVHGANRLGANSLLD
Sbjct 400 IPTNYHGEVI-TLRGDD---PDAVVPGLMAAGEAACASVHGANRLGANSLLD 447
> sce:YKL148C SDH1; Flavoprotein subunit of succinate dehydrogenase
(Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples the oxidation
of succinate to the transfer of electrons to ubiquinone
as part of the TCA cycle and the mitochondrial respiratory chain
(EC:1.3.5.1); K00234 succinate dehydrogenase (ubiquinone)
flavoprotein subunit [EC:1.3.5.1]
Length=640
Score = 251 bits (642), Expect = 7e-67, Method: Compositional matrix adjust.
Identities = 122/172 (70%), Positives = 139/172 (80%), Gaps = 3/172 (1%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHP+GI+ +GCLITEG RGEGG L NSEGE FM RYAPTAKDLA RDVVSR++T+EIR
Sbjct 284 VQFHPSGIYGSGCLITEGARGEGGFLVNSEGERFMERYAPTAKDLACRDVVSRAITMEIR 343
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG G +KDH +L L+HL PE L RLPGI+ETA IFAGVDVTK+PIP++PTVHYNMGG
Sbjct 344 EGRGVGKKKDHMYLQLSHLPPEVLKERLPGISETAAIFAGVDVTKEPIPIIPTVHYNMGG 403
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPT W E + + G E+ V+ GL A GEAAC SVHGANRLGANSLLD
Sbjct 404 IPTKWNGEALTIDEETG---EDKVIPGLMACGEAACVSVHGANRLGANSLLD 452
> cel:C03G5.1 sdha-1; Succinate DeHydrogenase complex subunit
A family member (sdha-1); K00234 succinate dehydrogenase (ubiquinone)
flavoprotein subunit [EC:1.3.5.1]
Length=646
Score = 249 bits (635), Expect = 4e-66, Method: Compositional matrix adjust.
Identities = 123/172 (71%), Positives = 137/172 (79%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ AGCLITEG RGEGG L NS GE FM RYAP AKDLASRDVVSRSMT+EI
Sbjct 274 VQFHPTGIYGAGCLITEGSRGEGGYLVNSAGERFMERYAPNAKDLASRDVVSRSMTVEIM 333
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG GP KDH +L L HL E L RLPGI+ETA IFAGVDVTK+PIPV+PTVHYNMGG
Sbjct 334 EGRGVGPDKDHIYLQLHHLPAEQLQQRLPGISETAMIFAGVDVTKEPIPVIPTVHYNMGG 393
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
+PTN++ +V++ KG + V+ GLYAAGE SVHGANRLGANSLLD
Sbjct 394 VPTNYKGQVLNYTPKKG----DEVVPGLYAAGECGAHSVHGANRLGANSLLD 441
> sce:YJL045W Minor succinate dehydrogenase isozyme; homologous
to Sdh1p, the major isozyme reponsible for the oxidation of
succinate and transfer of electrons to ubiquinone; induced
during the diauxic shift in a Cat8p-dependent manner (EC:1.3.5.1);
K00234 succinate dehydrogenase (ubiquinone) flavoprotein
subunit [EC:1.3.5.1]
Length=634
Score = 248 bits (633), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 122/172 (70%), Positives = 137/172 (79%), Gaps = 3/172 (1%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHP+GI+ +GCLITEG RGEGG L NSEGE FM RYAPTAKDLASRDVVSR++T+EIR
Sbjct 278 VQFHPSGIYGSGCLITEGARGEGGFLLNSEGERFMERYAPTAKDLASRDVVSRAITMEIR 337
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
GRG G KDH L L+HL PE L RLPGI+ETA +FAGVDVT++PIPVLPTVHYNMGG
Sbjct 338 AGRGVGKNKDHILLQLSHLPPEVLKERLPGISETAAVFAGVDVTQEPIPVLPTVHYNMGG 397
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPT W E + + G E+ V+ GL A GEAAC SVHGANRLGANSLLD
Sbjct 398 IPTKWTGEALTIDEETG---EDKVIPGLMACGEAACVSVHGANRLGANSLLD 446
> cel:C34B2.7 sdha-2; Succinate DeHydrogenase complex subunit
A family member (sdha-2); K00234 succinate dehydrogenase (ubiquinone)
flavoprotein subunit [EC:1.3.5.1]
Length=640
Score = 247 bits (630), Expect = 2e-65, Method: Compositional matrix adjust.
Identities = 121/172 (70%), Positives = 137/172 (79%), Gaps = 4/172 (2%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIR 60
VQFHPTGI+ GCLITEG RGEGG L NS+GE FM RYAP AKDLASRDVVSR+MT+EI
Sbjct 267 VQFHPTGIYGVGCLITEGSRGEGGYLVNSQGERFMERYAPNAKDLASRDVVSRAMTMEIN 326
Query 61 EGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
EGRG GP KDH +L L HL E L RLPGI+ETA+IFAGVDVTK+PIPV+PTVHYNMGG
Sbjct 327 EGRGVGPNKDHIYLQLHHLPAEQLQQRLPGISETAQIFAGVDVTKEPIPVIPTVHYNMGG 386
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
+PTN++ +V+ G + V+ GLYAAGE A SVHGANRLGANSLLD
Sbjct 387 VPTNYKGQVLDFTPEGG----DKVIPGLYAAGECAAHSVHGANRLGANSLLD 434
> eco:b0723 sdhA, ECK0712, JW0713; succinate dehydrogenase, flavoprotein
subunit (EC:1.3.99.1); K00239 succinate dehydrogenase
flavoprotein subunit [EC:1.3.99.1]
Length=588
Score = 208 bits (529), Expect = 8e-54, Method: Compositional matrix adjust.
Identities = 110/172 (63%), Positives = 122/172 (70%), Gaps = 5/172 (2%)
Query 2 QFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIEIRE 61
QFHPTGI AG L+TEGCRGEGG L N GE FM RYAP AKDLA RDVV+RS+ IEIRE
Sbjct 240 QFHPTGIAGAGVLVTEGCRGEGGYLLNKHGERFMERYAPNAKDLAGRDVVARSIMIEIRE 299
Query 62 GRGC-GPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGG 120
GRGC GP H L L HL E L SRLPGI E ++ FA VD K+PIPV+PT HY MGG
Sbjct 300 GRGCDGPWGPHAKLKLDHLGKEVLESRLPGILELSRTFAHVDPVKEPIPVIPTCHYMMGG 359
Query 121 IPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
IPT + + V KG E+ V+ GL+A GE AC SVHGANRLG NSLLD
Sbjct 360 IPTKVTGQAL-TVNEKG---EDVVVPGLFAVGEIACVSVHGANRLGGNSLLD 407
> eco:b4154 frdA, ECK4150, JW4115; fumarate reductase (anaerobic)
catalytic and NAD/flavoprotein subunit (EC:1.3.99.1); K00244
fumarate reductase flavoprotein subunit [EC:1.3.99.1]
Length=602
Score = 139 bits (351), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 83/182 (45%), Positives = 105/182 (57%), Gaps = 26/182 (14%)
Query 1 VQFHPTGIFPAGCLITEGCRGEGGILRNSEGEAFMARYA---------PTAK--DLASRD 49
VQ+HPTG+ +G L+TEGCRGEGGIL N G ++ Y P K +L RD
Sbjct 230 VQYHPTGLPGSGILMTEGCRGEGGILVNKNGYRYLQDYGMGPETPLGEPKNKYMELGPRD 289
Query 50 VVSRSMTIEIREGRGCG-PRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPI 108
VS++ E R+G PR D +LDL HL + LH RLP I E AK + GVD K+PI
Sbjct 290 KVSQAFWHEWRKGNTISTPRGDVVYLDLRHLGEKKLHERLPFICELAKAYVGVDPVKEPI 349
Query 109 PVLPTVHYNMGGIPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGAN 168
PV PT HY MGGI T+ +C E ++GL+A GE + +HGANRLG+N
Sbjct 350 PVRPTAHYTMGGIETDQ-----NC---------ETRIKGLFAVGECSSVGLHGANRLGSN 395
Query 169 SL 170
SL
Sbjct 396 SL 397
> ath:AT5G14760 AO; AO (L-ASPARTATE OXIDASE); L-aspartate oxidase/
electron carrier/ oxidoreductase; K00278 L-aspartate oxidase
[EC:1.4.3.16]
Length=651
Score = 105 bits (262), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 71/184 (38%), Positives = 96/184 (52%), Gaps = 33/184 (17%)
Query 1 VQFHPTGIFPAGC------------LITEGCRGEGGILRNSEGEAFMARYAPTAKDLASR 48
VQFHPT + G LITE RG+GGIL N E FM Y A +LA R
Sbjct 310 VQFHPTALADEGLPIKLQTARENAFLITEAVRGDGGILYNLGMERFMPVYDERA-ELAPR 368
Query 49 DVVSRSMTIEIREGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPI 108
DVV+RS+ ++++ + + LD++H E + + P I + G+D+T+QPI
Sbjct 369 DVVARSIDDQLKK-----RNEKYVLLDISHKPREKILAHFPNIASEC-LKHGLDITRQPI 422
Query 109 PVLPTVHYNMGGIPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGAN 168
PV+P HY GG+ G E VL GL+ AGE AC +HGANRL +N
Sbjct 423 PVVPAAHYMCGGVRA-------------GLQGETNVL-GLFVAGEVACTGLHGANRLASN 468
Query 169 SLLD 172
SLL+
Sbjct 469 SLLE 472
> eco:b2574 nadB, ECK2572, JW2558, nic, nicB; quinolinate synthase,
L-aspartate oxidase (B protein) subunit (EC:1.4.3.16);
K00278 L-aspartate oxidase [EC:1.4.3.16]
Length=540
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 61/175 (34%), Positives = 92/175 (52%), Gaps = 26/175 (14%)
Query 2 QFHPTGIF---PAGCLITEGCRGEGGILRNSEGEAFMARYAPTAKDLASRDVVSRSMTIE 58
QFHPT ++ L+TE RGEG L+ +G FM + +LA RD+V+R++ E
Sbjct 242 QFHPTALYHPQARNFLLTEALRGEGAYLKRPDGTRFMPDFDERG-ELAPRDIVARAIDHE 300
Query 59 I-REGRGCGPRKDHCHLDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYN 117
+ R G C LD++H + + P I E + G+D+T++P+P++P HY
Sbjct 301 MKRLGADC------MFLDISHKPADFIRQHFPMIYEKL-LGLGIDLTQEPVPIVPAAHYT 353
Query 118 MGGIPTNWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
GG+ V GR +EGLYA GE + +HGANR+ +NSLL+
Sbjct 354 CGGV----------MVDDHGRTD----VEGLYAIGEVSYTGLHGANRMASNSLLE 394
> sce:YEL047C Soluble fumarate reductase, required with isoenzyme
Osm1p for anaerobic growth; may interact with ribosomes,
based on co-purification experiments; authentic, non-tagged
protein is detected in purified mitochondria in high-throughput
studies (EC:1.3.1.6)
Length=470
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 65/223 (29%), Positives = 91/223 (40%), Gaps = 66/223 (29%)
Query 1 VQFHPTGIFPAG-------CLITEGCRGEGGILRNS-EGEAFMARYAPTAKDLASRDVVS 52
+Q HPTG L E RG GGIL N G F+ +L +RDVV+
Sbjct 246 IQVHPTGFIDPNDRSSSWKFLAAESLRGLGGILLNPITGRRFV-------NELTTRDVVT 298
Query 53 RSM--TIEIREGRGCGPRKDHCHLDL-THLDPETLHSRLPGIT----------------- 92
++ + R + + DL +LD + +T
Sbjct 299 AAIQKVCPQEDNRALLVMGEKMYTDLKNNLDFYMFKKLVQKLTLSQVVSEYNLPITVAQL 358
Query 93 ----ETAKIFA---------------GVDVTKQPI----PVLPTVHYNMGGIPTNWRSEV 129
+T F G DVT + + V P VH+ MGG N +++V
Sbjct 359 CEELQTYSSFTTKADPLGRTVILNEFGSDVTPETVVFIGEVTPVVHFTMGGARINVKAQV 418
Query 130 IHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
I GK +E +L+GLYAAGE + VHGANRLG +SLL+
Sbjct 419 I----GKN---DERLLKGLYAAGEVS-GGVHGANRLGGSSLLE 453
> sce:YJR051W OSM1; Fumarate reductase, catalyzes the reduction
of fumarate to succinate, required for the reoxidation of
intracellular NADH under anaerobic conditions; mutations cause
osmotic sensitivity
Length=501
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 64/229 (27%), Positives = 93/229 (40%), Gaps = 78/229 (34%)
Query 1 VQFHPTGIFPAG-------CLITEGCRGEGGILRN-SEGEAFMARYAPTAKDLASRDVVS 52
VQ HPTG L E RG GGIL + + G F +L++RD V
Sbjct 278 VQVHPTGFIDPNDRENNWKFLAAEALRGLGGILLHPTTGRRF-------TNELSTRDTV- 329
Query 53 RSMTIEIR------EGRGCGPRKDHCHLDLTH----------------------LDPETL 84
T+EI+ + R D + + T+ D +T
Sbjct 330 ---TMEIQSKCPKNDNRALLVMSDKVYENYTNNINFYMSKNLIKKVSINDLIRQYDLQTT 386
Query 85 HSRLPGITETAKIFAGV---DVTKQPI------------------PVLPTVHYNMGGIPT 123
S L +TE K ++ V D +P+ V P VH+ MGG+
Sbjct 387 ASEL--VTE-LKSYSDVNTKDTFDRPLIINAFDKDISTESTVYVGEVTPVVHFTMGGVKI 443
Query 124 NWRSEVIHCVKGKGRMAEECVLEGLYAAGEAACASVHGANRLGANSLLD 172
N +S+VI + +E + G++AAGE + VHGANRLG +SLL+
Sbjct 444 NEKSQVIK------KNSESVLSNGIFAAGEVS-GGVHGANRLGGSSLLE 485
> cel:F48E8.3 hypothetical protein
Length=493
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 43/82 (52%), Gaps = 17/82 (20%)
Query 91 ITETAKIFAGVDVTKQPIPVLPTVHYNMGGIPTNWRSEVIHCVKGKGRMAEECVLEGLYA 150
I+ T I+A + V+P +HY MGG+ + + VI GK + GL+A
Sbjct 406 ISPTEPIYAAI--------VVPAIHYTMGGLKIDEATRVIDE-HGK-------PIVGLFA 449
Query 151 AGEAACASVHGANRLGANSLLD 172
AGE VHG+NRL NSLL+
Sbjct 450 AGEV-TGGVHGSNRLAGNSLLE 470
> hsa:9829 DNAJC6, DJC6, KIAA0473, MGC129914, MGC129915, MGC48436;
DnaJ (Hsp40) homolog, subfamily C, member 6 (EC:3.1.3.48);
K09526 DnaJ homolog subfamily C member 6
Length=913
Score = 29.6 bits (65), Expect = 5.0, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 10/73 (13%)
Query 74 LDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGGIPT---NWRSE-- 128
LD HLD T+++ P TAK + V PI P++H N+ + NW +
Sbjct 98 LDSRHLDHYTVYNLSPKSYRTAKFHSRVSECSWPIRQAPSLH-NLFAVCRNMYNWLLQNP 156
Query 129 ----VIHCVKGKG 137
V+HC+ G+
Sbjct 157 KNVCVVHCLDGRA 169
> mmu:72685 Dnajc6, 2810027M23Rik, mKIAA0473; DnaJ (Hsp40) homolog,
subfamily C, member 6 (EC:3.1.3.48); K09526 DnaJ homolog
subfamily C member 6
Length=968
Score = 29.6 bits (65), Expect = 5.7, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 10/73 (13%)
Query 74 LDLTHLDPETLHSRLPGITETAKIFAGVDVTKQPIPVLPTVHYNMGGIPT---NWRSE-- 128
LD HLD T+++ P TAK + V PI P++H N+ + NW +
Sbjct 153 LDSRHLDHYTVYNLSPKSYRTAKFHSRVSECSWPIRQAPSLH-NLFAVCRNMYNWLLQNP 211
Query 129 ----VIHCVKGKG 137
V+HC+ G+
Sbjct 212 KNVCVVHCLDGRA 224
Lambda K H
0.320 0.138 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40