bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4730_orf1
Length=245
Score E
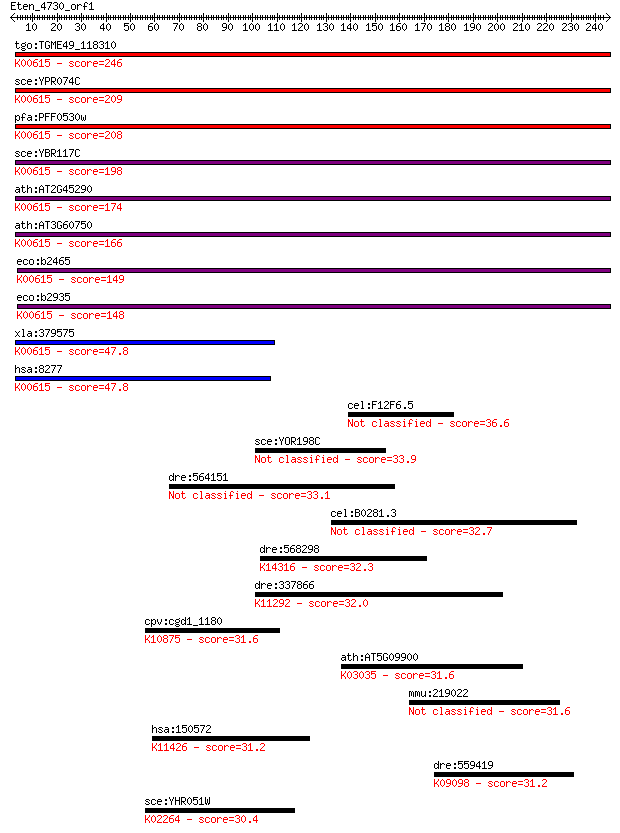
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_118310 transketolase, putative (EC:2.2.1.3); K00615... 246 6e-65
sce:YPR074C TKL1; Tkl1p (EC:2.2.1.1); K00615 transketolase [EC... 209 5e-54
pfa:PFF0530w PfTK; transketolase (EC:2.2.1.1); K00615 transket... 208 1e-53
sce:YBR117C TKL2; Tkl2p (EC:2.2.1.1); K00615 transketolase [EC... 198 1e-50
ath:AT2G45290 transketolase, putative (EC:2.2.1.1); K00615 tra... 174 4e-43
ath:AT3G60750 transketolase, putative (EC:2.2.1.1); K00615 tra... 166 7e-41
eco:b2465 tktB, ECK2460, JW2449; transketolase 2, thiamin-bind... 149 8e-36
eco:b2935 tktA, ECK2930, JW5478, tkt; transketolase 1, thiamin... 148 2e-35
xla:379575 tktl2, MGC69114; transketolase-like 2 (EC:2.2.1.1);... 47.8 3e-05
hsa:8277 TKTL1, TKR, TKT2; transketolase-like 1 (EC:2.2.1.1); ... 47.8 4e-05
cel:F12F6.5 srgp-1; Slit-Robo GAP homolog family member (srgp-1) 36.6 0.075
sce:YOR198C BFR1; Bfr1p 33.9 0.48
dre:564151 claspin homolog (Xenopus laevis)-like 33.1 0.97
cel:B0281.3 hypothetical protein 32.7 1.1
dre:568298 pom121, wu:fa16c07, wu:fl35g12; POM121 membrane gly... 32.3 1.5
dre:337866 supt6h, Spt6, fj42h11, wu:fj42h11; suppressor of Ty... 32.0 2.3
cpv:cgd1_1180 RAD54 like SWI/SNF2 ATpase ; K10875 DNA repair a... 31.6 2.7
ath:AT5G09900 EMB2107 (EMBRYO DEFECTIVE 2107); K03035 26S prot... 31.6 2.9
mmu:219022 Ttc5, 5930437N14, AW743060, MGC38092; tetratricopep... 31.6 3.0
hsa:150572 SMYD1, BOP, KMT3D, ZMYND18, ZMYND22; SET and MYND d... 31.2 3.3
dre:559419 npas3b, si:dkeyp-13e2.1; neuronal PAS domain protei... 31.2 3.4
sce:YHR051W COX6; Cox6p (EC:1.9.3.1); K02264 cytochrome c oxid... 30.4 5.4
> tgo:TGME49_118310 transketolase, putative (EC:2.2.1.3); K00615
transketolase [EC:2.2.1.1]
Length=699
Score = 246 bits (628), Expect = 6e-65, Method: Compositional matrix adjust.
Identities = 120/243 (49%), Positives = 163/243 (67%), Gaps = 5/243 (2%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
HLGL RL VLYD N ITIDG+ L+F+E V QRF AY WH V DG+ D++G++QA+
Sbjct 203 HLGLHRLTVLYDDNNITIDGELHLAFSEKVQQRFKAYDWHVDVVDDGNTDVAGLVQAMEN 262
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
AK TDKP+LI ++TTIG+ S GT KVHG+PLS ++++ +KE+ GL+P+E +PE+
Sbjct 263 AKKRTDKPTLICVRTTIGFLSSKAGTAKVHGSPLSEEELRAVKEQCGLSPDEKLQIPEEV 322
Query 123 RKFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAAA 182
++FYA V R + +AW LF +Y + YP E +E+ RMF ++ +V LK + + A
Sbjct 323 KQFYAQVQERGERSVEAWNALFERYKKEYPSEGEEIERMFSHKLRPEVLETLKTLGEKAK 382
Query 183 DTPASTRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGSYADRYIH 242
+ STR SG+ L IKDFMPELIGGSADLT SN T LK P+ + + + RYIH
Sbjct 383 EKADSTRAHSGRMLLGIKDFMPELIGGSADLTGSNCTDLK-----EPSFQVANRSGRYIH 437
Query 243 FGV 245
FGV
Sbjct 438 FGV 440
> sce:YPR074C TKL1; Tkl1p (EC:2.2.1.1); K00615 transketolase [EC:2.2.1.1]
Length=680
Score = 209 bits (533), Expect = 5e-54, Method: Compositional matrix adjust.
Identities = 116/251 (46%), Positives = 155/251 (61%), Gaps = 16/251 (6%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
HL LG LI +YD NKITIDG TS+SF E+V +R+ AYGW V++G+ D++GI +AI +
Sbjct 174 HLKLGNLIAIYDDNKITIDGATSISFDEDVAKRYEAYGWEVLYVENGNEDLAGIAKAIAQ 233
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
AK DKP+LI++ TTIG+GS + G+ VHGAPL ADD+KQLK K G NP++ F VP++
Sbjct 234 AKLSKDKPTLIKMTTTIGYGSLHAGSHSVHGAPLKADDVKQLKSKFGFNPDKSFVVPQEV 293
Query 123 RKFY------AGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKD 176
Y GV A NK W LF++Y + +P+ EL R ++ E+ L
Sbjct 294 YDHYQKTILKPGVEANNK-----WNKLFSEYQKKFPELGAELARRLSGQLPANWESKLPT 348
Query 177 MAKAAADTPASTRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGS- 235
A D+ +TR LS L + + +PELIGGSADLT SN TR K F P P GS
Sbjct 349 Y--TAKDSAVATRKLSETVLEDVYNQLPELIGGSADLTPSNLTRWKEALDFQP-PSSGSG 405
Query 236 -YADRYIHFGV 245
Y+ RYI +G+
Sbjct 406 NYSGRYIRYGI 416
> pfa:PFF0530w PfTK; transketolase (EC:2.2.1.1); K00615 transketolase
[EC:2.2.1.1]
Length=672
Score = 208 bits (530), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 106/243 (43%), Positives = 147/243 (60%), Gaps = 6/243 (2%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
HLGLGRLI+LYD NKITIDG+T LSFTEN+ ++F A W + V+DG++D IL I +
Sbjct 177 HLGLGRLILLYDDNKITIDGNTDLSFTENIEKKFEALNWEVRRVEDGNKDYKKILHEIEQ 236
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
K +P+LI ++T G+G++ EGT K HG L+ +D+K K GL+P + FH+ ++
Sbjct 237 GKKNLQQPTLIIVRTACGFGTKVEGTCKSHGLALNDEDLKNAKSFFGLDPQKKFHISDEV 296
Query 123 RKFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAAA 182
++FY V + K Y W+ +F + YP+ QE+ R F ++ + AL
Sbjct 297 KEFYKNVIQKKKENYIKWKNMFDDFSLKYPQVSQEIIRRFQNDLPNNWKDALPKY--TPK 354
Query 183 DTPASTRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGSYADRYIH 242
D P +TR LSG LN+I PELIGGSADL+ SN T LK E N SY ++YI
Sbjct 355 DAPGATRNLSGIVLNSINKIFPELIGGSADLSESNCTSLKEENDIKKN----SYGNKYIR 410
Query 243 FGV 245
FGV
Sbjct 411 FGV 413
> sce:YBR117C TKL2; Tkl2p (EC:2.2.1.1); K00615 transketolase [EC:2.2.1.1]
Length=681
Score = 198 bits (504), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 109/246 (44%), Positives = 149/246 (60%), Gaps = 6/246 (2%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
HL LG LI YDSN I+IDG TS SF E+VL+R+ AYGW V GD D+ I A+ +
Sbjct 174 HLQLGNLITFYDSNSISIDGKTSYSFDEDVLKRYEAYGWEVMEVDKGDDDMESISSALEK 233
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
AK DKP++I++ TTIG+GS +GT VHG+ L ADD+KQLK++ G +PN+ F VP++
Sbjct 234 AKLSKDKPTIIKVTTTIGFGSLQQGTAGVHGSALKADDVKQLKKRWGFDPNKSFVVPQEV 293
Query 123 RKFY-AGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAA 181
+Y V + + W +F +Y +P++ +EL R E+ E E K + K
Sbjct 294 YDYYKKTVVEPGQKLNEEWDRMFEEYKTKFPEKGKELQRRLNGELPEGWE---KHLPKFT 350
Query 182 ADTPA-STRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPN-PKFGSYADR 239
D A +TR S + L + +PELIGGSADLT SN TR +G F P + G+YA R
Sbjct 351 PDDDALATRKTSQQVLTNMVQVLPELIGGSADLTPSNLTRWEGAVDFQPPITQLGNYAGR 410
Query 240 YIHFGV 245
YI +GV
Sbjct 411 YIRYGV 416
> ath:AT2G45290 transketolase, putative (EC:2.2.1.1); K00615 transketolase
[EC:2.2.1.1]
Length=741
Score = 174 bits (440), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 105/245 (42%), Positives = 140/245 (57%), Gaps = 10/245 (4%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
H GLG+LI YD N I+IDGDT ++FTE+V +RF A GWH VK+G+ I AIR
Sbjct 250 HWGLGKLIAFYDDNHISIDGDTDIAFTESVDKRFEALGWHVIWVKNGNNGYDEIRAAIRE 309
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTE-KVHGAPLSADDIKQLKEKLGLNPNEDFHVPED 121
AKAVTDKP+LI++ TTIG+GS N+ VHGA L +++ + LG P E FHVPED
Sbjct 310 AKAVTDKPTLIKVTTTIGYGSPNKANSYSVHGAALGEKEVEATRNNLGW-PYEPFHVPED 368
Query 122 TRKFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAA 181
+ ++ A W F Y + YP+E EL + E+ E AL
Sbjct 369 VKSHWSRHTPEGAALEADWNAKFAAYEKKYPEEAAELKSIISGELPVGWEKALPTYTP-- 426
Query 182 ADTPA-STRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGSYADRY 240
D+P +TR LS + LNA+ +P +GGSADL +SN T LK AF N + + +R
Sbjct 427 -DSPGDATRNLSQQCLNALAKAVPGFLGGSADLASSNMTMLK---AFG-NFQKATPEERN 481
Query 241 IHFGV 245
+ FGV
Sbjct 482 LRFGV 486
> ath:AT3G60750 transketolase, putative (EC:2.2.1.1); K00615 transketolase
[EC:2.2.1.1]
Length=741
Score = 166 bits (420), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 99/245 (40%), Positives = 135/245 (55%), Gaps = 10/245 (4%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRR 62
H GLG+LI YD N I+IDGDT ++FTENV QRF A GWH VK+G+ I AI+
Sbjct 250 HWGLGKLIAFYDDNHISIDGDTEIAFTENVDQRFEALGWHVIWVKNGNTGYDEIRAAIKE 309
Query 63 AKAVTDKPSLIEIKTTIGWGSQNEGTE-KVHGAPLSADDIKQLKEKLGLNPNEDFHVPED 121
AK VTDKP+LI++ TTIG+GS N+ VHGA L +++ + LG P E F VP+D
Sbjct 310 AKTVTDKPTLIKVTTTIGYGSPNKANSYSVHGAALGEKEVEATRNNLGW-PYEPFQVPDD 368
Query 122 TRKFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAA 181
+ ++ W F Y + YP+E EL + E+ E AL
Sbjct 369 VKSHWSRHTPEGATLESDWSAKFAAYEKKYPEEASELKSIITGELPAGWEKALPTYTP-- 426
Query 182 ADTPA-STRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGSYADRY 240
++P +TR LS + LNA+ +P +GGSADL +SN T LK F + + +R
Sbjct 427 -ESPGDATRNLSQQCLNALAKVVPGFLGGSADLASSNMTLLKAFGDF----QKATPEERN 481
Query 241 IHFGV 245
+ FGV
Sbjct 482 LRFGV 486
> eco:b2465 tktB, ECK2460, JW2449; transketolase 2, thiamin-binding
(EC:2.2.1.1); K00615 transketolase [EC:2.2.1.1]
Length=667
Score = 149 bits (376), Expect = 8e-36, Method: Compositional matrix adjust.
Identities = 95/247 (38%), Positives = 133/247 (53%), Gaps = 15/247 (6%)
Query 4 LGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRRA 63
LGLG+LI YD N I+IDG+T FT++ +RF AY WH DG D + +AI A
Sbjct 172 LGLGKLIGFYDHNGISIDGETEGWFTDDTAKRFEAYHWHVIHEIDG-HDPQAVKEAILEA 230
Query 64 KAVTDKPSLIEIKTTIGWGSQNE-GTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
++V DKPSLI +T IG+GS N+ G E+ HGAPL +++ ++KLG + + F +P
Sbjct 231 QSVKDKPSLIICRTVIGFGSPNKAGKEEAHGAPLGEEEVALARQKLGWH-HPPFEIP--- 286
Query 123 RKFYAGVAARNKAE--YDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKA 180
++ Y AR K E +W F Y +A+P+ +E R + + E +
Sbjct 287 KEIYHAWDAREKGEKAQQSWNEKFAAYKKAHPQLAEEFTRRMSGGLPKDWEKTTQKYINE 346
Query 181 AADTPA--STRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGSYAD 238
PA +TR S LNA +PEL+GGSADL SN T KG + +P A
Sbjct 347 LQANPAKIATRKASQNTLNAYGPMLPELLGGSADLAPSNLTIWKGSVSLKEDP-----AG 401
Query 239 RYIHFGV 245
YIH+GV
Sbjct 402 NYIHYGV 408
> eco:b2935 tktA, ECK2930, JW5478, tkt; transketolase 1, thiamin-binding
(EC:2.2.1.1); K00615 transketolase [EC:2.2.1.1]
Length=663
Score = 148 bits (373), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 96/247 (38%), Positives = 128/247 (51%), Gaps = 15/247 (6%)
Query 4 LGLGRLIVLYDSNKITIDGDTSLSFTENVLQRFNAYGWHTQTVKDGDRDISGILQAIRRA 63
L LG+LI YD N I+IDG FT++ RF AYGWH DG D + I +A+ A
Sbjct 173 LKLGKLIAFYDDNGISIDGHVEGWFTDDTAMRFEAYGWHVIRDIDG-HDAASIKRAVEEA 231
Query 64 KAVTDKPSLIEIKTTIGWGSQNE-GTEKVHGAPLSADDIKQLKEKLGLNPNEDFHVPEDT 122
+AVTDKPSL+ KT IG+GS N+ GT HGAPL +I +E+LG F +P +
Sbjct 232 RAVTDKPSLLMCKTIIGFGSPNKAGTHDSHGAPLGDAEIALTREQLGWK-YAPFEIPSE- 289
Query 123 RKFYAGVAAR--NKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKA 180
YA A+ +A+ AW F Y +AYP+E E R E+ + K+
Sbjct 290 --IYAQWDAKEAGQAKESAWNEKFAAYAKAYPQEAAEFTRRMKGEMPSDFDAKAKEFIAK 347
Query 181 AADTPA--STRVLSGKALNAIKDFMPELIGGSADLTTSNETRLKGETAFSPNPKFGSYAD 238
PA ++R S A+ A +PE +GGSADL SN T G A + + A
Sbjct 348 LQANPAKIASRKASQNAIEAFGPLLPEFLGGSADLAPSNLTLWSGSKAINEDA-----AG 402
Query 239 RYIHFGV 245
YIH+GV
Sbjct 403 NYIHYGV 409
> xla:379575 tktl2, MGC69114; transketolase-like 2 (EC:2.2.1.1);
K00615 transketolase [EC:2.2.1.1]
Length=625
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 32/108 (29%), Positives = 55/108 (50%), Gaps = 4/108 (3%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENV-LQRFNAYGWHTQTVKDGDRDISGILQAIR 61
H L L+ ++D N++ L ++ ++R A+GW+T V DG D++ + A
Sbjct 172 HYHLDNLVAIFDVNRLGQSEAAPLQHQTDIYMKRCEAFGWNTYVV-DG-HDVAELCHAFW 229
Query 62 RAKAVTDKPSLIEIKTTIGWG-SQNEGTEKVHGAPLSADDIKQLKEKL 108
+A V DKP+ I KT G G S E + HG P+ D ++ + ++
Sbjct 230 QAAHVKDKPTAIIAKTFKGKGISGVENEDNWHGKPMPKDKVESIINEI 277
> hsa:8277 TKTL1, TKR, TKT2; transketolase-like 1 (EC:2.2.1.1);
K00615 transketolase [EC:2.2.1.1]
Length=590
Score = 47.8 bits (112), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 35/109 (32%), Positives = 53/109 (48%), Gaps = 7/109 (6%)
Query 3 HLGLGRLIVLYDSNKITIDGDTSLSFTENVLQR-FNAYGWHTQTVKDGDRDISGILQAIR 61
+ L L+ ++D N++ G N+ QR A+GW+T V DG RD+ + Q
Sbjct 137 YYSLDNLVAIFDVNRLGHSGALPAEHCINIYQRRCEAFGWNTYVV-DG-RDVEALCQVFW 194
Query 62 RAKAVTDKPSLIEIKTTIGWGSQN-EGTEKVHGAPLS---ADDIKQLKE 106
+A V KP+ + KT G G+ + E E H P+ AD I +L E
Sbjct 195 QASQVKHKPTAVVAKTFKGRGTPSIEDAESWHAKPMPRERADAIIKLIE 243
> cel:F12F6.5 srgp-1; Slit-Robo GAP homolog family member (srgp-1)
Length=1059
Score = 36.6 bits (83), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 28/43 (65%), Gaps = 1/43 (2%)
Query 139 AWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAA 181
W TL + E Y K++Q LG ++G++++ +ET +D+AK +
Sbjct 130 VWHTLVEQTKEEY-KKRQSLGELYGKQMTASIETRCEDLAKIS 171
> sce:YOR198C BFR1; Bfr1p
Length=470
Score = 33.9 bits (76), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 101 IKQLKEKL-GLNPNEDFHVPEDTRKFYAGVAARNKAEYDAWQTLFTKYGEAYPK 153
I QLKE+L GLNP + + E+ ++ + ++ + YD QTLF K Y K
Sbjct 167 INQLKEELNGLNPKDVSNQFEENQQKLNDIHSKTQGVYDKRQTLFNKRAALYKK 220
> dre:564151 claspin homolog (Xenopus laevis)-like
Length=920
Score = 33.1 bits (74), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 42/106 (39%), Gaps = 14/106 (13%)
Query 66 VTDKP--------SLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEK------LGLN 111
+TDKP S IE KTT G Q E V A +K K+K LGL+
Sbjct 65 ITDKPNPEDEEKNSPIEEKTTEGHPEQEEQQVSVDAAQTEPQVLKPRKDKLARLRELGLD 124
Query 112 PNEDFHVPEDTRKFYAGVAARNKAEYDAWQTLFTKYGEAYPKEKQE 157
P + D F A + A Q F K+ + P+ K+E
Sbjct 125 PPPVVKLSADEGAFVQLEAPQENQALKALQERFMKHFQPAPRPKRE 170
> cel:B0281.3 hypothetical protein
Length=329
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 48/119 (40%), Gaps = 20/119 (16%)
Query 132 RNKAEYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAAADTPASTRVL 191
+N E QTLF + GE + K+K ++ + + + E+V K+ D + RV
Sbjct 159 QNVEELKKAQTLFNENGELFQKKKNQITNFYSK-LREKVNEREKEALSELKDVARTARVN 217
Query 192 SGKA---LNAIKDFMPELIGGS----------------ADLTTSNETRLKGETAFSPNP 231
+ K+ L ++ ELI + A L SN + E F PNP
Sbjct 218 NTKSLLTLMQVRSIYDELISEAKELLKKTSATSFREVEAILNRSNAHIMSQEKIFEPNP 276
> dre:568298 pom121, wu:fa16c07, wu:fl35g12; POM121 membrane glycoprotein
(rat); K14316 nuclear pore complex protein Nup121
Length=1201
Score = 32.3 bits (72), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 25/76 (32%), Positives = 34/76 (44%), Gaps = 13/76 (17%)
Query 103 QLKEKLGLNPNEDFHVPEDTRKFYAG-------VAARNKAEYDAWQTLFTKYGEAYPKEK 155
QL E+LGLNP VP ++ G A RNK +T F A P ++
Sbjct 54 QLFERLGLNPRRGLSVPPALLRWLPGRTFSGVPAAGRNKIRKSDARTSF-----ASPSDR 108
Query 156 QELGRMFGRE-VSEQV 170
+G + RE +SE V
Sbjct 109 HFVGSYYRREQLSESV 124
> dre:337866 supt6h, Spt6, fj42h11, wu:fj42h11; suppressor of
Ty 6 homolog; K11292 transcription elongation factor SPT6
Length=1726
Score = 32.0 bits (71), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 33/106 (31%), Positives = 43/106 (40%), Gaps = 13/106 (12%)
Query 101 IKQLKEKLGLNPNEDFHVP--EDTRKFYAGVAARNKAEYDAWQTLFTKYGEAYPK---EK 155
I ++KE L N+ F VP RK Y + WQ + E + + K
Sbjct 344 IAKIKEALNFMRNQHFEVPFIAFYRKEYVEPELNINDLWKVWQ-----WDEKWTQLKTRK 398
Query 156 QELGRMFGREVSEQVETALKDMAKAAADTPASTRVLSGKALNAIKD 201
Q L R+F R S Q E D K AD STR L + +KD
Sbjct 399 QNLTRLFQRMQSYQFEQISADPDKPLAD---STRPLDTADMERLKD 441
> cpv:cgd1_1180 RAD54 like SWI/SNF2 ATpase ; K10875 DNA repair
and recombination protein RAD54 and RAD54-like protein [EC:3.6.4.-]
Length=877
Score = 31.6 bits (70), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 34/59 (57%), Gaps = 4/59 (6%)
Query 56 ILQAIRRAKAVTDKPSLIEIKTTIGWGSQNEGTEK----VHGAPLSADDIKQLKEKLGL 110
+L +I+ + + P+LI KT+ G+G EG+EK +HG +S + + K+++ +
Sbjct 432 VLSSIQSLMKLCNHPTLIRPKTSGGYGKGFEGSEKYLEMIHGRSVSGESGGEYKKRVTI 490
> ath:AT5G09900 EMB2107 (EMBRYO DEFECTIVE 2107); K03035 26S proteasome
regulatory subunit N5
Length=462
Score = 31.6 bits (70), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 5/75 (6%)
Query 136 EYDAWQTLFTKYGEAYPKEKQELGRMFGREVSEQVETALKDMAKAAADTPASTRVLSGKA 195
E W +L+ KY + + KEK +G G + E ++ + + K P +VL
Sbjct 303 EVIQWTSLWNKYKDEFEKEKSMIGGSLGDKAGEDLKLRIIEHVK----YPRCLKVLRKDN 358
Query 196 LN-AIKDFMPELIGG 209
L + FMPE GG
Sbjct 359 LKETCRAFMPEHGGG 373
> mmu:219022 Ttc5, 5930437N14, AW743060, MGC38092; tetratricopeptide
repeat domain 5
Length=440
Score = 31.6 bits (70), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 33/63 (52%), Gaps = 7/63 (11%)
Query 164 REVSEQVETALKDMAKA--AADTPASTRVLSGKALNAIKDFMPELIGGSADLTTSNETRL 221
++V E++E L+ M + +A A +L GKALN D+ PE A++ S +L
Sbjct 44 QDVQEEMEKTLQQMEEVLGSAQVEAQALMLKGKALNVTPDYSPE-----AEVLLSKAVKL 98
Query 222 KGE 224
+ E
Sbjct 99 EPE 101
> hsa:150572 SMYD1, BOP, KMT3D, ZMYND18, ZMYND22; SET and MYND
domain containing 1; K11426 SET and MYND domain-containing
protein
Length=490
Score = 31.2 bits (69), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 2/64 (3%)
Query 59 AIRRAKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNEDFHV 118
AI+R V ++ I + I W + EGT G +S DD++ E G +D V
Sbjct 92 AIKRYGKVPNEN--IRLAARIMWRVEREGTGLTEGCLVSVDDLQNHVEHFGEEEQKDLRV 149
Query 119 PEDT 122
DT
Sbjct 150 DVDT 153
> dre:559419 npas3b, si:dkeyp-13e2.1; neuronal PAS domain protein
3b; K09098 neuronal PAS domain-containing protein 1/3
Length=770
Score = 31.2 bits (69), Expect = 3.4, Method: Composition-based stats.
Identities = 21/59 (35%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Query 174 LKDMAKAAADTPASTRVLSGKALNAIKDFMPELIG--GSADLTTSNETRLKGETAFSPN 230
L DM P S R +A ++ +D P+ G G+ L + +ET LKG+ AFS N
Sbjct 415 LADMPLDLLQLPESLRAERLQASSSPRDMSPKTQGSTGTQPLKSRSETDLKGKEAFSHN 473
> sce:YHR051W COX6; Cox6p (EC:1.9.3.1); K02264 cytochrome c oxidase
subunit Va [EC:1.9.3.1]
Length=148
Score = 30.4 bits (67), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 33/61 (54%), Gaps = 5/61 (8%)
Query 56 ILQAIRRAKAVTDKPSLIEIKTTIGWGSQNEGTEKVHGAPLSADDIKQLKEKLGLNPNED 115
I +A+R A+ V D P+ I + + + +NE K + D++K ++++LG+ E+
Sbjct 88 IEKALRAARRVNDLPTAIRVFEALKYKVENEDQYKAY-----LDELKDVRQELGVPLKEE 142
Query 116 F 116
Sbjct 143 L 143
Lambda K H
0.314 0.132 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8636871780
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40