bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4723_orf1
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
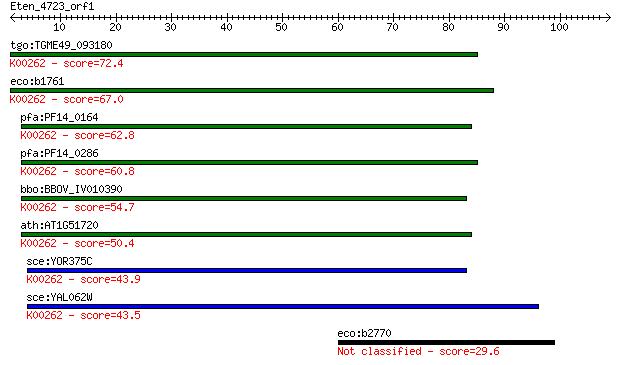
tgo:TGME49_093180 NADP-specific glutamate dehydrogenase, putat... 72.4 3e-13
eco:b1761 gdhA, ECK1759, JW1750; glutamate dehydrogenase, NADP... 67.0 1e-11
pfa:PF14_0164 NADP-specific glutamate dehydrogenase; K00262 gl... 62.8 3e-10
pfa:PF14_0286 glutamate dehydrogenase, putative (EC:1.4.1.3); ... 60.8 1e-09
bbo:BBOV_IV010390 23.m06170; glutamate dehydrogenase (EC:1.4.1... 54.7 7e-08
ath:AT1G51720 glutamate dehydrogenase, putative; K00262 glutam... 50.4 1e-06
sce:YOR375C GDH1, DHE4, URE1; Gdh1p (EC:1.4.1.4); K00262 gluta... 43.9 1e-04
sce:YAL062W GDH3, FUN51; Gdh3p (EC:1.4.1.4); K00262 glutamate ... 43.5 2e-04
eco:b2770 ygcR, ECK2765, JW5441; predicted flavoprotein 29.6 2.3
> tgo:TGME49_093180 NADP-specific glutamate dehydrogenase, putative
(EC:1.4.1.4); K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=489
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 40/85 (47%), Positives = 49/85 (57%), Gaps = 1/85 (1%)
Query 1 AREKLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEAKAADSCVRVRSFADGDSIV-F 59
A EKL +GAKV+T SDS G I +GF IQR+KE K S RV FA S V F
Sbjct 285 AGEKLATLGAKVMTFSDSSGYIVNEKGFPLGQIQRLKEMKETRSSTRVSEFAAKYSTVKF 344
Query 60 QNSETPWAVAADLAFPCAKENEVDD 84
+ W V D+AFPCA +NE+ +
Sbjct 345 VPDKRAWEVPCDIAFPCATQNEISE 369
> eco:b1761 gdhA, ECK1759, JW1750; glutamate dehydrogenase, NADP-specific
(EC:1.4.1.4); K00262 glutamate dehydrogenase (NADP+)
[EC:1.4.1.4]
Length=447
Score = 67.0 bits (162), Expect = 1e-11, Method: Composition-based stats.
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 1/87 (1%)
Query 1 AREKLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEAKAADSCVRVRSFADGDSIVFQ 60
A EK + GA+V+T SDS G + GF+KE + R+ E KA+ RV +A +V+
Sbjct 247 AIEKAMEFGARVITASDSSGTVVDESGFTKEKLARLIEIKASRDG-RVADYAKEFGLVYL 305
Query 61 NSETPWAVAADLAFPCAKENEVDDIAA 87
+ PW++ D+A PCA +NE+D AA
Sbjct 306 EGQQPWSLPVDIALPCATQNELDVDAA 332
> pfa:PF14_0164 NADP-specific glutamate dehydrogenase; K00262
glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=470
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 35/86 (40%), Positives = 50/86 (58%), Gaps = 10/86 (11%)
Query 3 EKLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQ---RIKEAKAADSCVRVRSFADGDSIV- 58
+KLL + KVLTLSDS G +Y GF+ E+++ +KE K R++ + + S
Sbjct 269 QKLLHLNVKVLTLSDSNGYVYEPNGFTHENLEFLIDLKEEKKG----RIKEYLNHSSTAK 324
Query 59 -FQNSETPWAVAADLAFPCAKENEVD 83
F N E PW V LAFPCA +NE++
Sbjct 325 YFPN-EKPWGVPCTLAFPCATQNEIN 349
> pfa:PF14_0286 glutamate dehydrogenase, putative (EC:1.4.1.3);
K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=510
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 32/84 (38%), Positives = 50/84 (59%), Gaps = 4/84 (4%)
Query 3 EKLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEAKAADSCVRVRSFADGDSIV--FQ 60
EKL+ GA VLT+SDS G I GF+KE + I + K + +R++ + F+
Sbjct 310 EKLIEKGAIVLTMSDSNGYILEPNGFTKEQLNYIMDIK-NNQRLRLKEYLKYSKTAKYFE 368
Query 61 NSETPWAVAADLAFPCAKENEVDD 84
N + PW + D+AFPCA +NE+++
Sbjct 369 N-QKPWNIPCDIAFPCATQNEINE 391
> bbo:BBOV_IV010390 23.m06170; glutamate dehydrogenase (EC:1.4.1.4);
K00262 glutamate dehydrogenase (NADP+) [EC:1.4.1.4]
Length=455
Score = 54.7 bits (130), Expect = 7e-08, Method: Composition-based stats.
Identities = 29/81 (35%), Positives = 46/81 (56%), Gaps = 2/81 (2%)
Query 3 EKLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEAKAADSCVRVRSFAD-GDSIVFQN 61
EKL+ +GA +T+SDS G I +G + E ++ I K S R+ + + + F
Sbjct 255 EKLIELGAVPITMSDSSGYIIEPEGITLEGLRYIMAFKNPHS-RRISEYLNYSKTATFHP 313
Query 62 SETPWAVAADLAFPCAKENEV 82
+ PW +AD+AFPCA +NE+
Sbjct 314 GDKPWGESADIAFPCATQNEI 334
> ath:AT1G51720 glutamate dehydrogenase, putative; K00262 glutamate
dehydrogenase (NADP+) [EC:1.4.1.4]
Length=637
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 44/81 (54%), Gaps = 1/81 (1%)
Query 3 EKLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEAKAADSCVRVRSFADGDSIVFQNS 62
EKL+A GA +T+SDS+G + GF + +++ K+ +R S + F +
Sbjct 434 EKLIACGAHPVTVSDSKGYLVDDDGFDYMKLAFLRDIKSQQRSLRDYSKTYARAKYF-DE 492
Query 63 ETPWAVAADLAFPCAKENEVD 83
PW D+AFPCA +NEVD
Sbjct 493 LKPWNERCDVAFPCASQNEVD 513
> sce:YOR375C GDH1, DHE4, URE1; Gdh1p (EC:1.4.1.4); K00262 glutamate
dehydrogenase (NADP+) [EC:1.4.1.4]
Length=454
Score = 43.9 bits (102), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 41/85 (48%), Gaps = 6/85 (7%)
Query 4 KLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEA----KAADSCVRVRSFADGDSIVF 59
K++ +G V++LSDS+G I G + E + I A K+ + V S + + +
Sbjct 235 KVIELGGTVVSLSDSKGCIISETGITSEQVADISSAKVNFKSLEQIVNEYSTFSENKVQY 294
Query 60 QNSETPWAVA--ADLAFPCAKENEV 82
PW D+A PCA +NEV
Sbjct 295 IAGARPWTHVQKVDIALPCATQNEV 319
> sce:YAL062W GDH3, FUN51; Gdh3p (EC:1.4.1.4); K00262 glutamate
dehydrogenase (NADP+) [EC:1.4.1.4]
Length=457
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 52/105 (49%), Gaps = 18/105 (17%)
Query 4 KLLAMGAKVLTLSDSEGMIYCSQGFSKEDIQRIKEAKAADSCVRVRSFAD--GDSIVFQN 61
K++ +G V++LSDS+G I G + E I I AK +R +S + + F
Sbjct 236 KVIELGGIVVSLSDSKGCIISETGITSEQIHDIASAK-----IRFKSLEEIVDEYSTFSE 290
Query 62 SET-------PWAVAA--DLAFPCAKENEV--DDIAAAMSSRAEF 95
S+ PW + D+A PCA +NEV D+ A ++S +F
Sbjct 291 SKMKYVAGARPWTHVSNVDIALPCATQNEVSGDEAKALVASGVKF 335
> eco:b2770 ygcR, ECK2765, JW5441; predicted flavoprotein
Length=259
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Query 60 QNSETPWAVAADLAFPCAKENE---VDDIAAAMSSRAEFGTR 98
QN +TP+ +A L +PC + E +D + + R E G R
Sbjct 130 QNGQTPFLLAEMLGWPCFTQVERFTLDALFITLEQRTEHGLR 171
Lambda K H
0.316 0.128 0.355
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003209916
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40