bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4710_orf1
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
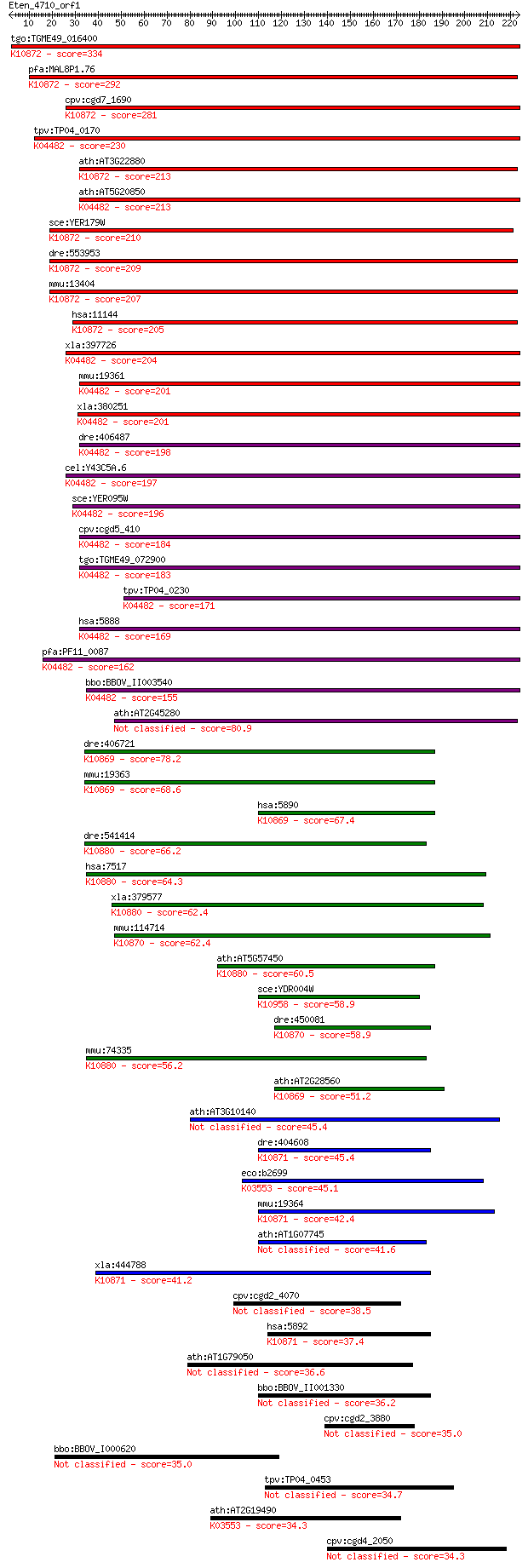
tgo:TGME49_016400 meiotic recombination protein DMC1-like prot... 334 1e-91
pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;... 292 5e-79
cpv:cgd7_1690 meiotic recombination protein DMC1-like protein ... 281 2e-75
tpv:TP04_0170 meiotic recombination protein DMC1; K04482 DNA r... 230 4e-60
ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1); AT... 213 3e-55
ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-depende... 213 4e-55
sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination pr... 210 4e-54
dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor... 209 7e-54
mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage sup... 207 3e-53
hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473, d... 205 1e-52
xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad... 204 2e-52
mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cer... 201 1e-51
xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,... 201 2e-51
dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA h... 198 1e-50
cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-... 197 4e-50
sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51 196 6e-50
cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51 184 2e-46
tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8); K... 183 5e-46
tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair prot... 171 1e-42
hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA... 169 7e-42
pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein ... 162 8e-40
bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair ... 155 1e-37
ath:AT2G45280 ATRAD51C; ATP binding / damaged DNA binding / pr... 80.9 3e-15
dre:406721 rad51l1, wu:fd07f04, zgc:56581; RAD51-like 1 (S. ce... 78.2 2e-14
mmu:19363 Rad51l1, AI553500, R51H2, Rad51b, mREC2; RAD51-like ... 68.6 2e-11
hsa:5890 RAD51L1, MGC34245, R51H2, RAD51B, REC2, hREC2; RAD51-... 67.4 3e-11
dre:541414 xrcc3, im:7142103, si:dkey-11b8.1, zgc:101608; X-ra... 66.2 8e-11
hsa:7517 XRCC3, CMM6; X-ray repair complementing defective rep... 64.3 3e-10
xla:379577 xrcc3, MGC69118; X-ray repair complementing defecti... 62.4 1e-09
mmu:114714 Rad51c, R51H3, Rad51l2; RAD51 homolog c (S. cerevis... 62.4 1e-09
ath:AT5G57450 XRCC3; XRCC3; ATP binding / damaged DNA binding ... 60.5 4e-09
sce:YDR004W RAD57; Protein that stimulates strand exchange by ... 58.9 1e-08
dre:450081 rad51c, zgc:101596; rad51 homolog C (S. cerevisiae)... 58.9 1e-08
mmu:74335 Xrcc3, 4432412E01Rik, AI182522, AW537713; X-ray repa... 56.2 8e-08
ath:AT2G28560 RAD51B; RAD51B; recombinase; K10869 RAD51-like p... 51.2 3e-06
ath:AT3G10140 RECA3; RECA3 (recA homolog 3); ATP binding / DNA... 45.4 1e-04
dre:404608 MGC77165; zgc:77165; K10871 RAD51-like protein 3 45.4
eco:b2699 recA, ECK2694, JW2669, lexB, recH, rnmB, srf, tif, u... 45.1 2e-04
mmu:19364 Rad51l3, DKFZp586D0122, R51H3, Rad51d, Trad-d2, Trad... 42.4 0.001
ath:AT1G07745 RAD51D; RAD51D (ARABIDOPSIS HOMOLOG OF RAD51 D);... 41.6 0.002
xla:444788 rad51l3, MGC82048; RAD51-like 3; K10871 RAD51-like ... 41.2 0.003
cpv:cgd2_4070 hypothetical protein 38.5 0.021
hsa:5892 RAD51L3, R51H3, RAD51D, TRAD; RAD51-like 3 (S. cerevi... 37.4 0.039
ath:AT1G79050 DNA repair protein recA 36.6 0.078
bbo:BBOV_II001330 18.m06101; hypothetical protein 36.2 0.087
cpv:cgd2_3880 hypothetical protein 35.0 0.21
bbo:BBOV_I000620 16.m00943; hypothetical protein 35.0 0.23
tpv:TP04_0453 hypothetical protein 34.7 0.31
ath:AT2G19490 recA family protein; K03553 recombination protei... 34.3 0.36
cpv:cgd4_2050 hypothetical protein 34.3 0.40
> tgo:TGME49_016400 meiotic recombination protein DMC1-like protein,
putative (EC:2.7.11.1); K10872 meiotic recombination
protein DMC1
Length=349
Score = 334 bits (857), Expect = 1e-91, Method: Compositional matrix adjust.
Identities = 164/222 (73%), Positives = 189/222 (85%), Gaps = 2/222 (0%)
Query 2 ESKSARKHVGTLPASCASDLNEGNDEHFICIDKLQSVGINAADIAKLRGSGYCTVLSVIQ 61
ESKSARKH+ + P S ++ ND F IDKLQ+ GINAADI KL+ +GYCTVLS++Q
Sbjct 6 ESKSARKHISSTPHLDVS-ADDANDT-FQSIDKLQAAGINAADINKLKQAGYCTVLSIVQ 63
Query 62 TTKKDLALVKGLSEAKVEKIVEAAMKLELCNSFISGGELIARRAKVLKITTGSPQLDLLL 121
TTKK+L LVKG+SEAKVEKIVEAA KL +CN+FI+G EL+ +R +V+KITTGS QLD LL
Sbjct 64 TTKKELCLVKGISEAKVEKIVEAAAKLGMCNAFITGVELVQKRGRVIKITTGSDQLDQLL 123
Query 122 GGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQ 181
GGG ETMSITE+FGENRCGKTQ+CHT+CVTAQLPR+M GGCGKVCYIDTE TFRPEKI
Sbjct 124 GGGFETMSITELFGENRCGKTQLCHTVCVTAQLPRDMKGGCGKVCYIDTEGTFRPEKIQG 183
Query 182 IAERFGLDGAGCLDNIMYARAYTTEHMHQLFTVAAAKMSEEK 223
IAERFGLDG G LDNIMYARA+TTEHMHQL T+AAAKM EE+
Sbjct 184 IAERFGLDGDGVLDNIMYARAFTTEHMHQLLTLAAAKMCEER 225
> pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;
K10872 meiotic recombination protein DMC1
Length=347
Score = 292 bits (748), Expect = 5e-79, Method: Compositional matrix adjust.
Identities = 146/222 (65%), Positives = 178/222 (80%), Gaps = 9/222 (4%)
Query 10 VGTLPAS-CASDL-------NEGNDEH-FICIDKLQSVGINAADIAKLRGSGYCTVLSVI 60
+ TLPAS AS + +EG EH F I+KLQ +GINAADI KL+GSGYCT+LS+I
Sbjct 1 MATLPASKSASKVALTSEVEDEGIKEHTFQEIEKLQDLGINAADINKLKGSGYCTILSLI 60
Query 61 QTTKKDLALVKGLSEAKVEKIVEAAMKLELCNSFISGGELIARRAKVLKITTGSPQLDLL 120
QTTKK+L VKG+SEAKV+KI+E A K+E C+SFI+ EL+ +R+KVLKITTGS D
Sbjct 61 QTTKKELCNVKGISEAKVDKILEVASKIENCSSFITANELVQKRSKVLKITTGSTVFDQT 120
Query 121 LGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKIC 180
LGGGIE+M ITE+FGENRCGKTQ+CHTL VTAQLP+ + GG GKVCYIDTE TFRPEK+C
Sbjct 121 LGGGIESMCITELFGENRCGKTQVCHTLAVTAQLPKSLNGGNGKVCYIDTEGTFRPEKVC 180
Query 181 QIAERFGLDGAGCLDNIMYARAYTTEHMHQLFTVAAAKMSEE 222
+IAER+GLDG LDNI+YARA+T EH++QL ++AAKM EE
Sbjct 181 KIAERYGLDGEAVLDNILYARAFTHEHLYQLLAISAAKMCEE 222
> cpv:cgd7_1690 meiotic recombination protein DMC1-like protein
; K10872 meiotic recombination protein DMC1
Length=342
Score = 281 bits (718), Expect = 2e-75, Method: Compositional matrix adjust.
Identities = 137/198 (69%), Positives = 160/198 (80%), Gaps = 0/198 (0%)
Query 26 DEHFICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAA 85
+E F+ IDKLQS GIN ADI KL+ +G CTVLS+IQ TKK+L +KGLSEAKVEKIVEAA
Sbjct 21 EEAFVEIDKLQSAGINVADINKLKTAGLCTVLSIIQATKKELCNIKGLSEAKVEKIVEAA 80
Query 86 MKLELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQIC 145
KL+ +SF SG E+++RR +L+ITTGS Q D +L GG E+M ITEIFGENRCGKTQIC
Sbjct 81 QKLDQSSSFQSGSEVMSRRQNILRITTGSEQFDKMLMGGFESMCITEIFGENRCGKTQIC 140
Query 146 HTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTT 205
HTLCV AQLP EM GG GKVC+IDTE TFRPE+I +IAERFG+ G LDNIMYARAYT
Sbjct 141 HTLCVAAQLPLEMNGGNGKVCFIDTEGTFRPERIVKIAERFGVQGDVALDNIMYARAYTH 200
Query 206 EHMHQLFTVAAAKMSEEK 223
EH++QL + AA KM EEK
Sbjct 201 EHLNQLISAAAGKMIEEK 218
> tpv:TP04_0170 meiotic recombination protein DMC1; K04482 DNA
repair protein RAD51
Length=346
Score = 230 bits (586), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 120/214 (56%), Positives = 152/214 (71%), Gaps = 6/214 (2%)
Query 12 TLPASCASDLNEGNDEHFICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVK 71
++ +S D+++ + F I++L+ +GIN DI KL+ +G CTVL VIQTTKKDL +K
Sbjct 13 SIQSSTNEDISDTILKPFQPIERLEELGINVTDINKLKAAGICTVLGVIQTTKKDLCNIK 72
Query 72 GLSEAKVEKIVEAAMKLELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSIT 131
GL+E KV+KI + A KLE+ NSFIS EL R +LKI TGS L+ LL GGIETMSIT
Sbjct 73 GLTELKVDKISDCASKLEVTNSFISASELYKIRKSILKINTGSEMLNRLLNGGIETMSIT 132
Query 132 EIFGENRCGKTQICHTLCVTAQL--PREMGGGCGKVCYIDTEATFRPEKICQIAERFGLD 189
E+FGENR GKTQICHT+ VT+Q+ P E KVCYIDTE TFRPEKI +I ERF LD
Sbjct 133 ELFGENRTGKTQICHTISVTSQIINPTE----PFKVCYIDTENTFRPEKIEKICERFDLD 188
Query 190 GAGCLDNIMYARAYTTEHMHQLFTVAAAKMSEEK 223
LDNI+Y++AYT EH+ QL + +KM EE+
Sbjct 189 PMITLDNILYSKAYTNEHLLQLISNITSKMVEER 222
> ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1);
ATP binding / DNA binding / DNA-dependent ATPase/ damaged DNA
binding / nucleoside-triphosphatase/ nucleotide binding /
protein binding; K10872 meiotic recombination protein DMC1
Length=344
Score = 213 bits (543), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 112/191 (58%), Positives = 137/191 (71%), Gaps = 1/191 (0%)
Query 32 IDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELC 91
IDKL + GINA D+ KL+ +G T ++ TKK+L +KGLSEAKV+KI EAA K+ +
Sbjct 31 IDKLIAQGINAGDVKKLQEAGIHTCNGLMMHTKKNLTGIKGLSEAKVDKICEAAEKI-VN 89
Query 92 NSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVT 151
+++G + + +R V+KITTG LD LLGGGIET +ITE FGE R GKTQ+ HTLCVT
Sbjct 90 FGYMTGSDALIKRKSVVKITTGCQALDDLLGGGIETSAITEAFGEFRSGKTQLAHTLCVT 149
Query 152 AQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQL 211
QLP M GG GKV YIDTE TFRP++I IAERFG+D LDNI+YARAYT EH + L
Sbjct 150 TQLPTNMKGGNGKVAYIDTEGTFRPDRIVPIAERFGMDPGAVLDNIIYARAYTYEHQYNL 209
Query 212 FTVAAAKMSEE 222
AAKMSEE
Sbjct 210 LLGLAAKMSEE 220
> ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-dependent
ATPase/ damaged DNA binding / nucleoside-triphosphatase/
nucleotide binding / protein binding / sequence-specific DNA
binding; K04482 DNA repair protein RAD51
Length=342
Score = 213 bits (542), Expect = 4e-55, Method: Compositional matrix adjust.
Identities = 105/192 (54%), Positives = 142/192 (73%), Gaps = 1/192 (0%)
Query 32 IDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELC 91
+++LQ+ GI + D+ KLR +G CTV V T +KDL +KG+S+AKV+KIVEAA KL +
Sbjct 28 VEQLQAAGIASVDVKKLRDAGLCTVEGVAYTPRKDLLQIKGISDAKVDKIVEAASKL-VP 86
Query 92 NSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVT 151
F S +L A+R ++++IT+GS +LD +L GGIET SITE++GE R GKTQ+CHTLCVT
Sbjct 87 LGFTSASQLHAQRQEIIQITSGSRELDKVLEGGIETGSITELYGEFRSGKTQLCHTLCVT 146
Query 152 AQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQL 211
QLP + GGG GK YID E TFRP+++ QIA+RFGL+GA L+N+ YARAY T+H +L
Sbjct 147 CQLPMDQGGGEGKAMYIDAEGTFRPQRLLQIADRFGLNGADVLENVAYARAYNTDHQSRL 206
Query 212 FTVAAAKMSEEK 223
AA+ M E +
Sbjct 207 LLEAASMMIETR 218
> sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination
protein DMC1
Length=334
Score = 210 bits (534), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 107/202 (52%), Positives = 141/202 (69%), Gaps = 1/202 (0%)
Query 19 SDLNEGNDEHFICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKV 78
++++ ++ + +D+LQ+ GINA+D+ KL+ G TV +V+ TT++ L +KGLSE KV
Sbjct 6 TEIDSDTAKNILSVDELQNYGINASDLQKLKSGGIYTVNTVLSTTRRHLCKIKGLSEVKV 65
Query 79 EKIVEAAMKLELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENR 138
EKI EAA K+ + FI + R +V ++TGS QLD +LGGGI TMSITE+FGE R
Sbjct 66 EKIKEAAGKI-IQVGFIPATVQLDIRQRVYSLSTGSKQLDSILGGGIMTMSITEVFGEFR 124
Query 139 CGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIM 198
CGKTQ+ HTLCVT QLPREMGGG GKV YIDTE TFRPE+I QIAE + LD CL N+
Sbjct 125 CGKTQMSHTLCVTTQLPREMGGGEGKVAYIDTEGTFRPERIKQIAEGYELDPESCLANVS 184
Query 199 YARAYTTEHMHQLFTVAAAKMS 220
YARA +EH +L ++S
Sbjct 185 YARALNSEHQMELVEQLGEELS 206
> dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=342
Score = 209 bits (532), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 110/204 (53%), Positives = 136/204 (66%), Gaps = 1/204 (0%)
Query 19 SDLNEGNDEHFICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKV 78
S + + F I+ LQ GIN ADI KL+ G CTV + TT++ L +KGLSEAKV
Sbjct 13 SGYQDDEESFFQDIELLQKHGINVADIKKLKSVGICTVKGIQMTTRRALCNIKGLSEAKV 72
Query 79 EKIVEAAMKLELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENR 138
+KI EAA KL C F + E +R +V ITTGS + D LLGGG+E+M+ITE FGE R
Sbjct 73 DKIKEAAGKLLTCG-FQTASEYCIKRKQVFHITTGSLEFDKLLGGGVESMAITEAFGEFR 131
Query 139 CGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIM 198
GKTQ+ HTLCVTAQLP E G GKV +IDTE TFRPE++ IA+RF +D LDN++
Sbjct 132 TGKTQLSHTLCVTAQLPGEYGYTGGKVIFIDTENTFRPERLKDIADRFNVDHEAVLDNVL 191
Query 199 YARAYTTEHMHQLFTVAAAKMSEE 222
YARAYT+EH +L AAK EE
Sbjct 192 YARAYTSEHQMELLDFVAAKFHEE 215
> mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=340
Score = 207 bits (527), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 109/204 (53%), Positives = 135/204 (66%), Gaps = 1/204 (0%)
Query 19 SDLNEGNDEHFICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKV 78
S + + F ID LQ GIN ADI KL+ G CT+ + TT++ L VKGLSEAKV
Sbjct 11 SGFQDDEESLFQDIDLLQKHGINMADIKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKV 70
Query 79 EKIVEAAMKLELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENR 138
EKI EAA KL + F++ + RR V ITTGS + D LLGGGIE+M+ITE FGE R
Sbjct 71 EKIKEAANKL-IEPGFLTAFQYSERRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFR 129
Query 139 CGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIM 198
GKTQ+ HTLCVTAQLP G GK+ +IDTE TFRP+++ IA+RF +D LDN++
Sbjct 130 TGKTQLSHTLCVTAQLPGTGGYSGGKIIFIDTENTFRPDRLRDIADRFNVDHEAVLDNVL 189
Query 199 YARAYTTEHMHQLFTVAAAKMSEE 222
YARAYT+EH +L AAK EE
Sbjct 190 YARAYTSEHQMELLDYVAAKFHEE 213
> hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473,
dJ199H16.1; DMC1 dosage suppressor of mck1 homolog, meiosis-specific
homologous recombination (yeast); K10872 meiotic recombination
protein DMC1
Length=340
Score = 205 bits (521), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 107/194 (55%), Positives = 132/194 (68%), Gaps = 1/194 (0%)
Query 29 FICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKL 88
F ID LQ GIN ADI KL+ G CT+ + TT++ L VKGLSEAKV+KI EAA KL
Sbjct 21 FQDIDLLQKHGINVADIKKLKSVGICTIKGIQMTTRRALCNVKGLSEAKVDKIKEAANKL 80
Query 89 ELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTL 148
+ F++ E +R V ITTGS + D LLGGGIE+M+ITE FGE R GKTQ+ HTL
Sbjct 81 -IEPGFLTAFEYSEKRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTL 139
Query 149 CVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHM 208
CVTAQLP G GK+ +IDTE TFRP+++ IA+RF +D LDN++YARAYT+EH
Sbjct 140 CVTAQLPGAGGYPGGKIIFIDTENTFRPDRLRDIADRFNVDHDAVLDNVLYARAYTSEHQ 199
Query 209 HQLFTVAAAKMSEE 222
+L AAK EE
Sbjct 200 MELLDYVAAKFHEE 213
> xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad51;
RAD51 homolog (RecA homolog); K04482 DNA repair protein
RAD51
Length=336
Score = 204 bits (519), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 104/200 (52%), Positives = 139/200 (69%), Gaps = 3/200 (1%)
Query 26 DEHF--ICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVE 83
+EHF I +L+ GINA D+ KL +G+ TV +V KK+L +KG+SEAK EKI+
Sbjct 14 EEHFGPQAISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELLNIKGISEAKAEKILA 73
Query 84 AAMKLELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQ 143
A KL + F + E RR+++++I+TGS +LD LL GG+ET SITE+FGE R GKTQ
Sbjct 74 EAAKL-VPMGFTTATEFHQRRSEIIQISTGSKELDKLLQGGVETGSITEMFGEFRTGKTQ 132
Query 144 ICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAY 203
+CHTL VT QLP + GGG GK YIDTE TFRPE++ +AER+GL G+ LDN+ YARA+
Sbjct 133 LCHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAF 192
Query 204 TTEHMHQLFTVAAAKMSEEK 223
T+H QL A+A M+E +
Sbjct 193 NTDHQTQLLYQASAMMAESR 212
> mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cerevisiae);
K04482 DNA repair protein RAD51
Length=339
Score = 201 bits (512), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 103/192 (53%), Positives = 133/192 (69%), Gaps = 1/192 (0%)
Query 32 IDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELC 91
I +L+ GINA D+ KL +GY TV +V KK+L +KG+SEAK +KI+ A KL +
Sbjct 25 ISRLEQCGINANDVKKLEEAGYHTVEAVAYAPKKELINIKGISEAKADKILTEAAKL-VP 83
Query 92 NSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVT 151
F + E RR+++++ITTGS +LD LL GGIET SITE+FGE R GKTQICHTL VT
Sbjct 84 MGFTTATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQICHTLAVT 143
Query 152 AQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQL 211
QLP + GGG GK YIDTE TFRPE++ +AER+GL G+ LDN+ YAR + T+H QL
Sbjct 144 CQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARGFNTDHQTQL 203
Query 212 FTVAAAKMSEEK 223
A+A M E +
Sbjct 204 LYQASAMMVESR 215
> xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,
reca, xrad51; RAD51 homolog (RecA homolog); K04482 DNA repair
protein RAD51
Length=336
Score = 201 bits (511), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 102/193 (52%), Positives = 134/193 (69%), Gaps = 1/193 (0%)
Query 31 CIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLEL 90
I +L+ GINA D+ KL +G+ TV +V KK+L +KG+SEAK EKI+ A KL +
Sbjct 21 AITRLEQCGINANDVKKLEDAGFHTVEAVAYAPKKELLNIKGISEAKAEKILAEAAKL-V 79
Query 91 CNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCV 150
F + E RR+++++I TGS +LD LL GGIET SITE+FGE R GKTQ+CHTL V
Sbjct 80 PMGFTTATEFHQRRSEIIQIGTGSKELDKLLQGGIETGSITEMFGEFRTGKTQLCHTLAV 139
Query 151 TAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQ 210
T QLP + GGG GK YIDTE TFRPE++ +AER+GL G+ LDN+ YARA+ T+H Q
Sbjct 140 TCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFNTDHQTQ 199
Query 211 LFTVAAAKMSEEK 223
L A+A M+E +
Sbjct 200 LLYQASAMMAESR 212
> dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA
homolog, E. coli) (S. cerevisiae); K04482 DNA repair protein
RAD51
Length=338
Score = 198 bits (503), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 99/192 (51%), Positives = 135/192 (70%), Gaps = 1/192 (0%)
Query 32 IDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELC 91
+ +L+ GI+++DI KL G+ TV +V KK+L +KG+SEAK +KI+ A K+ +
Sbjct 24 VSRLEQSGISSSDIKKLEDGGFHTVEAVAYAPKKELLNIKGISEAKADKILTEAAKM-VP 82
Query 92 NSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVT 151
F + E RRA++++I+TGS +LD LL GGIET SITE+FGE R GKTQ+CHTL VT
Sbjct 83 MGFTTATEFHQRRAEIIQISTGSKELDKLLQGGIETGSITEMFGEFRTGKTQLCHTLAVT 142
Query 152 AQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQL 211
QLP + GGG GK YIDTE TFRPE++ +AER+GL G+ LDN+ YARA+ T+H QL
Sbjct 143 CQLPIDQGGGEGKAMYIDTEGTFRPERLLAVAERYGLVGSDVLDNVAYARAFNTDHQTQL 202
Query 212 FTVAAAKMSEEK 223
A+A M+E +
Sbjct 203 LYQASAMMTESR 214
> cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-related
family member (rad-51); K04482 DNA repair protein
RAD51
Length=395
Score = 197 bits (500), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 105/198 (53%), Positives = 142/198 (71%), Gaps = 1/198 (0%)
Query 26 DEHFICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAA 85
DE+F IDKL+S GI++ DI+KL+ +GY T S+ TT+++L VKG+S+ K EKI++ A
Sbjct 73 DENFTVIDKLESSGISSGDISKLKEAGYYTYESLAFTTRRELRNVKGISDQKAEKIMKEA 132
Query 86 MKLELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQIC 145
MK + F +G E+ +R+++++I TGS LD LLGGGIET SITE++GE R GKTQ+C
Sbjct 133 MKF-VQMGFTTGAEVHVKRSQLVQIRTGSASLDRLLGGGIETGSITEVYGEYRTGKTQLC 191
Query 146 HTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTT 205
H+L V QLP +MGGG GK YIDT ATFRPE+I IA+R+ +D A L+NI ARAY +
Sbjct 192 HSLAVLCQLPIDMGGGEGKCMYIDTNATFRPERIIAIAQRYNMDSAHVLENIAVARAYNS 251
Query 206 EHMHQLFTVAAAKMSEEK 223
EH+ L A A MSE +
Sbjct 252 EHLMALIIRAGAMMSESR 269
> sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51
Length=400
Score = 196 bits (498), Expect = 6e-50, Method: Compositional matrix adjust.
Identities = 98/195 (50%), Positives = 133/195 (68%), Gaps = 1/195 (0%)
Query 29 FICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKL 88
F+ I+KLQ GI AD+ KLR SG T +V +KDL +KG+SEAK +K++ A +L
Sbjct 80 FVPIEKLQVNGITMADVKKLRESGLHTAEAVAYAPRKDLLEIKGISEAKADKLLNEAARL 139
Query 89 ELCNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTL 148
+ F++ + RR++++ +TTGS LD LLGGG+ET SITE+FGE R GK+Q+CHTL
Sbjct 140 -VPMGFVTAADFHMRRSELICLTTGSKNLDTLLGGGVETGSITELFGEFRTGKSQLCHTL 198
Query 149 CVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHM 208
VT Q+P ++GGG GK YIDTE TFRP ++ IA+RFGLD L+N+ YARAY +H
Sbjct 199 AVTCQIPLDIGGGEGKCLYIDTEGTFRPVRLVSIAQRFGLDPDDALNNVAYARAYNADHQ 258
Query 209 HQLFTVAAAKMSEEK 223
+L AA MSE +
Sbjct 259 LRLLDAAAQMMSESR 273
> cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51
Length=347
Score = 184 bits (467), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 100/192 (52%), Positives = 123/192 (64%), Gaps = 1/192 (0%)
Query 32 IDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELC 91
++ L G+ D+ LR +GY T+ + KK L VKG+SE K +KI A +L +
Sbjct 31 LEHLLPSGLTKRDLEILRENGYHTIECLAYAPKKALLSVKGISEQKCDKIKSACKEL-VA 89
Query 92 NSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVT 151
F SG E + R ++K TTGS QLD LL GGIET SITEIFGE R GKTQ+CHTL VT
Sbjct 90 MGFCSGTEYLEARTNLIKFTTGSSQLDRLLQGGIETGSITEIFGEFRTGKTQLCHTLAVT 149
Query 152 AQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQL 211
QLP E GG GK +IDTE TFRPE+I QIA+RF L+ + CLDNI YAR + TEH L
Sbjct 150 CQLPVEHKGGEGKCLWIDTEGTFRPERIVQIADRFNLNASDCLDNIAYARGFNTEHQMDL 209
Query 212 FTVAAAKMSEEK 223
A A M+E +
Sbjct 210 LQSAVAMMTESR 221
> tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8);
K04482 DNA repair protein RAD51
Length=354
Score = 183 bits (464), Expect = 5e-46, Method: Compositional matrix adjust.
Identities = 98/193 (50%), Positives = 127/193 (65%), Gaps = 3/193 (1%)
Query 32 IDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELC 91
++ L + G D+ L+ +GY TV + K+L VKGLSE KVEK+ +A+ ELC
Sbjct 38 LEHLLAKGFTKRDLELLKDAGYQTVECIAFAPVKNLVAVKGLSEQKVEKLKKASK--ELC 95
Query 92 N-SFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCV 150
N F S E + R +++ TTGS QLD LL GGIET ++TE+FGE R GKTQ+CHTL V
Sbjct 96 NLGFCSAQEYLEARENLIRFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAV 155
Query 151 TAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQ 210
T QLP E GG GK +IDTE TFRPE+I IA+RFGL+ CLDN+ YARAY +H +
Sbjct 156 TCQLPIEQAGGEGKCLWIDTEGTFRPERIVSIAKRFGLNANDCLDNVAYARAYNCDHQME 215
Query 211 LFTVAAAKMSEEK 223
L A+A M+E +
Sbjct 216 LLMEASAMMAESR 228
> tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair protein
RAD51
Length=343
Score = 171 bits (434), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 91/174 (52%), Positives = 121/174 (69%), Gaps = 3/174 (1%)
Query 51 SGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCN-SFISGGELIARRAKVLK 109
+GY T+ V +K+L ++KGLSE KV KI +AA + ELC+ F SG + + R ++K
Sbjct 47 AGYSTLECVAYAPQKNLLVIKGLSEQKVLKI-KAACR-ELCHLGFCSGQDYLEARGNLIK 104
Query 110 ITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYID 169
TTGS QLD LL GG+ET SITEI GE + GK+Q+CHTL VT QLP E GG GK ++D
Sbjct 105 FTTGSAQLDKLLQGGVETGSITEIIGEFKTGKSQLCHTLAVTCQLPVEQSGGEGKCLWVD 164
Query 170 TEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQLFTVAAAKMSEEK 223
+E TFRPE+I IA+RFGL + CLDN+ YARAY T+H +L A+A M++ +
Sbjct 165 SEGTFRPERIVSIAKRFGLSPSDCLDNVAYARAYNTDHQLELLVEASAMMAQTR 218
> hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA;
RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae); K04482
DNA repair protein RAD51
Length=340
Score = 169 bits (428), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 95/197 (48%), Positives = 126/197 (63%), Gaps = 10/197 (5%)
Query 32 IDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIV---EAAMKL 88
I +L+ GINA D+ KL +G+ TV +V KK+L +KG+SEAK +KI+ + +L
Sbjct 25 ISRLEQCGINANDVKKLEEAGFHTVEAVAYAPKKELINIKGISEAKADKILTESRSVARL 84
Query 89 ELCNSFISGGELIARRAKVLKITTGSPQLDLLLG--GGIETMSITEIFGENRCGKTQICH 146
E CNS I L+ ++ + ++G GGIET SITE+FGE R GKTQICH
Sbjct 85 E-CNSVI----LVYCTLRLSGSSDSPASASRVVGTTGGIETGSITEMFGEFRTGKTQICH 139
Query 147 TLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTE 206
TL VT QLP + GGG GK YIDTE TFRPE++ +AER+GL G+ LDN+ YARA+ T+
Sbjct 140 TLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERLLAVAERYGLSGSDVLDNVAYARAFNTD 199
Query 207 HMHQLFTVAAAKMSEEK 223
H QL A+A M E +
Sbjct 200 HQTQLLYQASAMMVESR 216
> pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein
RAD51
Length=350
Score = 162 bits (410), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 95/213 (44%), Positives = 130/213 (61%), Gaps = 7/213 (3%)
Query 16 SCASDLNEGNDEHF----ICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVK 71
S +S ++E +E + I++L + G D+ L+ G TV V + L +K
Sbjct 15 SNSSTIDEIEEEQLYTGPLKIEQLLAKGFVKRDLELLKEGGLQTVECVAYAPMRTLCAIK 74
Query 72 GLSEAKVEKIVEAAMKLELCNS-FISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSI 130
G+SE K EK+ +A ELCNS F + + R ++K TTGS QLD LL GGIET I
Sbjct 75 GISEQKAEKLKKACK--ELCNSGFCNAIDYHDARQNLIKFTTGSKQLDALLKGGIETGGI 132
Query 131 TEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDG 190
TE+FGE R GK+Q+CHTL +T QLP E GG GK +IDTE TFRPE+I IA+R+GL
Sbjct 133 TELFGEFRTGKSQLCHTLAITCQLPIEQSGGEGKCLWIDTEGTFRPERIVAIAKRYGLHP 192
Query 191 AGCLDNIMYARAYTTEHMHQLFTVAAAKMSEEK 223
CL+NI YA+AY +H +L A+A M++ +
Sbjct 193 TDCLNNIAYAKAYNCDHQTELLIDASAMMADAR 225
> bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair
protein RAD51
Length=346
Score = 155 bits (391), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 89/190 (46%), Positives = 114/190 (60%), Gaps = 3/190 (1%)
Query 35 LQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCNSF 94
L S G DI L+ +GY T+ S+ Q K L VKGLSE KV KI E ELC
Sbjct 31 LLSKGFLQRDIDVLKAAGYVTLDSIAQVASKTLLEVKGLSEQKVAKIKEIVK--ELCPPD 88
Query 95 I-SGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQ 153
I + E + R ++K TTGS LD LL GGIE+ SITEI G+ GKTQ+CHTL +T+Q
Sbjct 89 ICTAAEYLECRLNLIKFTTGSTALDALLQGGIESGSITEIIGDFSTGKTQLCHTLAITSQ 148
Query 154 LPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQLFT 213
LP E GG GK +IDT+ +FRPE++ IA RFGL A C+ NI+Y + TE +
Sbjct 149 LPIEQNGGEGKCLWIDTQNSFRPERLGPIANRFGLSHAECVANIVYVKVSNTEQQFDMLV 208
Query 214 VAAAKMSEEK 223
AA M++ +
Sbjct 209 EAAHYMAQSR 218
> ath:AT2G45280 ATRAD51C; ATP binding / damaged DNA binding /
protein binding / recombinase/ single-stranded DNA binding
Length=363
Score = 80.9 bits (198), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 64/183 (34%), Positives = 93/183 (50%), Gaps = 19/183 (10%)
Query 47 KLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCN---SFISGG----E 99
KL +GY + S+ + DLA ++E + +I++ A + CN S I+G +
Sbjct 36 KLISAGYTCLSSIASVSSSDLARDANITEEEAFEILKLANQ-SCCNGSRSLINGAKNAWD 94
Query 100 LIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMG 159
++ + +ITT LD +LGGGI +TEI G GKTQI L V Q+PRE G
Sbjct 95 MLHEEESLPRITTSCSDLDNILGGGISCRDVTEIGGVPGIGKTQIGIQLSVNVQIPRECG 154
Query 160 GGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQLFTVAAAKM 219
G GK YIDTE +F E+ QIAE C++++ YT +MH+ F +M
Sbjct 155 GLGGKAIYIDTEGSFMVERALQIAE-------ACVEDM---EEYTG-YMHKHFQANQVQM 203
Query 220 SEE 222
E
Sbjct 204 KPE 206
> dre:406721 rad51l1, wu:fd07f04, zgc:56581; RAD51-like 1 (S.
cerevisiae); K10869 RAD51-like protein 1
Length=373
Score = 78.2 bits (191), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 52/157 (33%), Positives = 85/157 (54%), Gaps = 9/157 (5%)
Query 34 KLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEA---KVEKIVEAAMKLEL 90
KL+ G++A +L+ T V+ T+ +L+ + GLS ++++V A
Sbjct 5 KLRRSGVSADLCERLKRHQLETCQDVLSVTQVELSRLAGLSYPAALNLQRLVSKA----- 59
Query 91 CNSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCV 150
C + + +R + L +T P LD LL GG+ ++TE+ G + CGKTQ+C L V
Sbjct 60 CAPAVITALDLWKRKEELCFSTSLPALDRLLHGGLPRGALTEVTGPSGCGKTQLCMMLSV 119
Query 151 TAQLPREMGGGCGKVCYIDTEATFRPEKICQIAE-RF 186
A LP+ +GG V YIDTE+ F E++ ++A+ RF
Sbjct 120 LATLPKSLGGLDSGVIYIDTESAFSAERLVEMAQSRF 156
> mmu:19363 Rad51l1, AI553500, R51H2, Rad51b, mREC2; RAD51-like
1 (S. cerevisiae); K10869 RAD51-like protein 1
Length=350
Score = 68.6 bits (166), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 54/157 (34%), Positives = 76/157 (48%), Gaps = 5/157 (3%)
Query 34 KLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCNS 93
KL+ VG++ +L + + +L V GLS V +++ K
Sbjct 5 KLRRVGLSPELCDRLSRYQIVNCQHFLSLSPLELMKVTGLSYRGVHELLHTVSK-ACAPQ 63
Query 94 FISGGELIARRAKVLK---ITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCV 150
+ EL RR+ L ++T LD L GG+ S+TEI G CGKTQ C + V
Sbjct 64 MQTAYELKTRRSAHLSPAFLSTTLCALDEALHGGVPCGSLTEITGPPGCGKTQFCIMMSV 123
Query 151 TAQLPREMGGGCGKVCYIDTEATFRPEKICQIAE-RF 186
A LP +GG G V YIDTE+ F E++ +IAE RF
Sbjct 124 LATLPTSLGGLEGAVVYIDTESAFTAERLVEIAESRF 160
> hsa:5890 RAD51L1, MGC34245, R51H2, RAD51B, REC2, hREC2; RAD51-like
1 (S. cerevisiae); K10869 RAD51-like protein 1
Length=350
Score = 67.4 bits (163), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/78 (47%), Positives = 47/78 (60%), Gaps = 1/78 (1%)
Query 110 ITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYID 169
++T LD L GG+ S+TEI G CGKTQ C + + A LP MGG G V YID
Sbjct 83 LSTTLSALDEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYID 142
Query 170 TEATFRPEKICQIAE-RF 186
TE+ F E++ +IAE RF
Sbjct 143 TESAFSAERLVEIAESRF 160
> dre:541414 xrcc3, im:7142103, si:dkey-11b8.1, zgc:101608; X-ray
repair complementing defective repair in Chinese hamster
cells 3; K10880 DNA-repair protein XRCC3
Length=352
Score = 66.2 bits (160), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 44/153 (28%), Positives = 79/153 (51%), Gaps = 4/153 (2%)
Query 34 KLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCNS 93
+ + + +N IA ++ + + V+ + DL + LS+A V+++ +A +
Sbjct 2 EWEQLELNPRIIAAVKKGNFRSAKEVLCVSGPDLQRLTRLSKADVQRLHQAVAASVRKSK 61
Query 94 FISGGELIARRAKVL----KITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLC 149
++ +LI VL +++ P LD L+ GG+ ITE+ GE+ GKTQ C LC
Sbjct 62 PVTALQLIQGECPVLEPGHRLSFACPVLDGLMRGGLPLRGITELAGESAAGKTQFCLQLC 121
Query 150 VTAQLPREMGGGCGKVCYIDTEATFRPEKICQI 182
++ Q P+E GG YI TE +F +++ Q+
Sbjct 122 LSVQYPQEHGGLNSGAVYICTEDSFPIKRLRQL 154
> hsa:7517 XRCC3, CMM6; X-ray repair complementing defective repair
in Chinese hamster cells 3; K10880 DNA-repair protein
XRCC3
Length=346
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 49/182 (26%), Positives = 84/182 (46%), Gaps = 8/182 (4%)
Query 35 LQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCNSF 94
L + +N IA ++ + +V V+ + DL + LS +V ++ A +S
Sbjct 3 LDLLDLNPRIIAAIKKAKLKSVKEVLHFSGPDLKRLTNLSSPEVWHLLRTASLHLRGSSI 62
Query 95 ISGGELIARRAKVL----KITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCV 150
++ +L ++ + +++ G P LD LL GG+ ITE+ G + GKTQ+ LC+
Sbjct 63 LTALQLHQQKERFPTQHQRLSLGCPVLDALLRGGLPLDGITELAGRSSAGKTQLALQLCL 122
Query 151 TAQLPREMGGGCGKVCYIDTEATFRPEKICQI---AERFGLDGAG-CLDNIMYARAYTTE 206
Q PR+ GG YI TE F +++ Q+ R D G L + + E
Sbjct 123 AVQFPRQHGGLEAGAVYICTEDAFPHKRLQQLMAQQPRLRTDVPGELLQKLRFGSQIFIE 182
Query 207 HM 208
H+
Sbjct 183 HV 184
> xla:379577 xrcc3, MGC69118; X-ray repair complementing defective
repair in Chinese hamster cells 3; K10880 DNA-repair protein
XRCC3
Length=350
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 51/170 (30%), Positives = 81/170 (47%), Gaps = 13/170 (7%)
Query 46 AKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCNSFISGGELIARRA 105
A ++ +G LSV + K + LS A V + +A + N ++ ++ + +A
Sbjct 19 ANIKSAGSILTLSVPELQK-----LANLSVADVRHLQKAVSAVLRKNLGVTALQMYSEKA 73
Query 106 KVL----KITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGG 161
K K++ G LD L GGI + ITEI GE+ GKTQI LC++ Q P E GG
Sbjct 74 KFPSQHQKLSLGCKVLDNFLRGGIPLVGITEIAGESSAGKTQIGLQLCLSVQYPVEYGGL 133
Query 162 CGKVCYIDTEATFRPEKICQIAE---RFGLD-GAGCLDNIMYARAYTTEH 207
YI TE F +++ Q+ + + D + + NI + + EH
Sbjct 134 ASGAVYICTEDAFPSKRLQQLIKSQHKLRSDVPSDVIKNIRFGDSIFVEH 183
> mmu:114714 Rad51c, R51H3, Rad51l2; RAD51 homolog c (S. cerevisiae);
K10870 RAD51-like protein 2
Length=366
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 49/175 (28%), Positives = 78/175 (44%), Gaps = 18/175 (10%)
Query 47 KLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCNSFISGG-------- 98
KL +G+ T V++ +L+ G+S+ + + ++ + L N G
Sbjct 17 KLVAAGFQTAEDVLEVKPSELSKEVGISKEEALETLQILRRECLTNKPRCAGTSVANEKC 76
Query 99 ---ELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLP 155
EL+ + I T LD +LGGGI M TE+ G GKTQ+C L V Q+P
Sbjct 77 TALELLEQEHTQGFIITFCSALDNILGGGIPLMKTTEVCGVPGVGKTQLCMQLAVDVQIP 136
Query 156 REMGGGCGKVCYIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEHMHQ 210
GG G+ +IDTE +F +++ +A C+ ++ TE HQ
Sbjct 137 ECFGGVAGEAVFIDTEGSFMVDRVVSLA-------TACIQHLHLIAGTHTEEEHQ 184
> ath:AT5G57450 XRCC3; XRCC3; ATP binding / damaged DNA binding
/ protein binding / single-stranded DNA binding; K10880 DNA-repair
protein XRCC3
Length=304
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 37/95 (38%), Positives = 49/95 (51%), Gaps = 0/95 (0%)
Query 92 NSFISGGELIARRAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVT 151
N I L+ R K+TTG LD L GGI S+TEI E+ CGKTQ+C L +
Sbjct 3 NGKIKPENLLRRSPTNRKLTTGCEILDGCLRGGISCDSLTEIVAESGCGKTQLCLQLSLC 62
Query 152 AQLPREMGGGCGKVCYIDTEATFRPEKICQIAERF 186
QLP GG G Y+ +E F ++ Q++ F
Sbjct 63 TQLPISHGGLNGSSLYLHSEFPFPFRRLHQLSHTF 97
> sce:YDR004W RAD57; Protein that stimulates strand exchange by
stabilizing the binding of Rad51p to single-stranded DNA;
involved in the recombinational repair of double-strand breaks
in DNA during vegetative growth and meiosis; forms heterodimer
with Rad55p; K10958 DNA repair protein RAD57
Length=460
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 31/70 (44%), Positives = 40/70 (57%), Gaps = 0/70 (0%)
Query 110 ITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYID 169
TT +D LLGGGI T ITEIFGE+ GK+Q+ L ++ QL GG GK YI
Sbjct 100 FTTADVAMDELLGGGIFTHGITEIFGESSTGKSQLLMQLALSVQLSEPAGGLGGKCVYIT 159
Query 170 TEATFRPEKI 179
TE +++
Sbjct 160 TEGDLPTQRL 169
> dre:450081 rad51c, zgc:101596; rad51 homolog C (S. cerevisiae);
K10870 RAD51-like protein 2
Length=362
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 30/68 (44%), Positives = 39/68 (57%), Gaps = 0/68 (0%)
Query 117 LDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRP 176
LD +GGG+ TEI G GKTQ+C L V Q+P GG GK YIDTE +F
Sbjct 93 LDDAIGGGVPVGKTTEICGAPGVGKTQLCMQLAVDVQIPVFFGGLGGKALYIDTEGSFLV 152
Query 177 EKICQIAE 184
+++ +AE
Sbjct 153 QRVADMAE 160
> mmu:74335 Xrcc3, 4432412E01Rik, AI182522, AW537713; X-ray repair
complementing defective repair in Chinese hamster cells
3; K10880 DNA-repair protein XRCC3
Length=349
Score = 56.2 bits (134), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 41/152 (26%), Positives = 70/152 (46%), Gaps = 4/152 (2%)
Query 35 LQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCNSF 94
L + +N A ++ +V ++ + DL + GL V+ ++ AA +
Sbjct 3 LDQLDLNPRITAAVKRGRLKSVKEILCYSGPDLQRLTGLPSHDVQCLLRAASLHLRGSRV 62
Query 95 ISGGELIARRAKVL----KITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCV 150
+S L ++ +++ G P LD LGGG+ IT + G + GKTQ+ LC+
Sbjct 63 LSALHLFQQKESFPEQHQRLSLGCPVLDQFLGGGLPLEGITGLAGCSSAGKTQLALQLCL 122
Query 151 TAQLPREMGGGCGKVCYIDTEATFRPEKICQI 182
Q PR+ GG YI TE F +++ Q+
Sbjct 123 AVQFPRQYGGLEAGAVYICTEDAFPSKRLWQL 154
> ath:AT2G28560 RAD51B; RAD51B; recombinase; K10869 RAD51-like
protein 1
Length=370
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query 117 LDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRP 176
LD L GGI +TE+ G GK+Q C L ++A P GG G+V YID E+ F
Sbjct 91 LDDTLCGGIPFGVLTELVGPPGIGKSQFCMKLALSASFPVAYGGLDGRVIYIDVESKFSS 150
Query 177 EKICQI-----AERFGLDG 190
++ ++ E F L G
Sbjct 151 RRVIEMGLESFPEVFHLKG 169
> ath:AT3G10140 RECA3; RECA3 (recA homolog 3); ATP binding / DNA
binding / DNA-dependent ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=389
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 41/145 (28%), Positives = 66/145 (45%), Gaps = 24/145 (16%)
Query 80 KIVEAAMKLELCNSFISGG---------ELIARRAKVLKITTGSPQLDLLLG-GGIETMS 129
K+ E L L S +SG + R+ +V I+TGS LDL LG GG+
Sbjct 58 KVAEKDTALHLALSQLSGDFDKDSKLSLQRFYRKRRVSVISTGSLNLDLALGVGGLPKGR 117
Query 130 ITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPEKICQIAERFGLD 189
+ E++G+ GKT + + AQ ++GG C Y+D E P +AE G++
Sbjct 118 MVEVYGKEASGKTTLALHIIKEAQ---KLGGYC---AYLDAENAMDP----SLAESIGVN 167
Query 190 GAGCLDNIMYARAYTTEHMHQLFTV 214
+ ++ +R + E M + V
Sbjct 168 ----TEELLISRPSSAEKMLNIVDV 188
> dre:404608 MGC77165; zgc:77165; K10871 RAD51-like protein 3
Length=327
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/77 (36%), Positives = 42/77 (54%), Gaps = 9/77 (11%)
Query 110 ITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCV--TAQLPREMGGGCGKVCY 167
++TGSP LD LL G+ T ITE+ G GKTQ+C ++ V + QL + V Y
Sbjct 82 LSTGSPSLDKLLDSGLYTGEITELTGSPGSGKTQVCFSVAVNISHQLKQ-------TVVY 134
Query 168 IDTEATFRPEKICQIAE 184
IDT+ ++ Q+ +
Sbjct 135 IDTKGGMCANRLLQMLQ 151
> eco:b2699 recA, ECK2694, JW2669, lexB, recH, rnmB, srf, tif,
umuB, umuR, zab; DNA strand exchange and recombination protein
with protease and nuclease activity; K03553 recombination
protein RecA
Length=353
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 39/107 (36%), Positives = 52/107 (48%), Gaps = 17/107 (15%)
Query 103 RRAKVLKITTGSPQLDLLLG-GGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGG 161
R V I+TGS LD+ LG GG+ I EI+G GKT + TL V A RE
Sbjct 34 RSMDVETISTGSLSLDIALGAGGLPMGRIVEIYGPESSGKTTL--TLQVIAAAQRE---- 87
Query 162 CGKVC-YIDTEATFRPEKICQIAERFGLDGAGCLDNIMYARAYTTEH 207
GK C +ID E P A + G+D +DN++ ++ T E
Sbjct 88 -GKTCAFIDAEHALDP----IYARKLGVD----IDNLLCSQPDTGEQ 125
> mmu:19364 Rad51l3, DKFZp586D0122, R51H3, Rad51d, Trad-d2, Trad-d3,
Trad-d4, Trad-d6, Trad-d7; RAD51-like 3 (S. cerevisiae);
K10871 RAD51-like protein 3
Length=329
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 47/106 (44%), Gaps = 8/106 (7%)
Query 110 ITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYID 169
++TG LD LL G+ T +TEI G GKTQ+C LCV A + + V Y+D
Sbjct 82 LSTGIGSLDKLLDAGLYTGEVTEIVGGPGSGKTQVC--LCVAANVAHSL---QQNVLYVD 136
Query 170 TEATFRPEKICQIAERFGLD---GAGCLDNIMYARAYTTEHMHQLF 212
+ ++ Q+ + D A L I R++ M +
Sbjct 137 SNGGMTASRLLQLLQARTQDEEKQASALQRIQVVRSFDIFRMLDML 182
> ath:AT1G07745 RAD51D; RAD51D (ARABIDOPSIS HOMOLOG OF RAD51 D);
ATP binding / DNA binding / DNA-dependent ATPase
Length=304
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 5/73 (6%)
Query 110 ITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYID 169
++TG + D LL GG +TE+ G + GKTQ C + G+V Y+D
Sbjct 71 LSTGDKETDSLLQGGFREGQLTELVGPSSSGKTQFCMQAAASVA-----ENHLGRVLYLD 125
Query 170 TEATFRPEKICQI 182
T +F +I Q
Sbjct 126 TGNSFSARRIAQF 138
> xla:444788 rad51l3, MGC82048; RAD51-like 3; K10871 RAD51-like
protein 3
Length=324
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 34/147 (23%), Positives = 67/147 (45%), Gaps = 7/147 (4%)
Query 39 GINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEKIVEAAMKLELCNSFISGG 98
G++ +A L+ + TV+ ++ + ++LA LS + + + F SG
Sbjct 11 GLSTGIVAALKANNVKTVIDLVASDLEELARKCSLSYKTLMAVRRVLLAQYSAFPF-SGA 69
Query 99 ELIAR-RAKVLKITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPRE 157
++ ++ + T + +LD+LL G+ T +TEI G GKTQ+C ++ V +
Sbjct 70 DVYEELKSSTAILPTANRKLDILLDSGLYTGEVTEIAGAAGSGKTQMCQSIAVNVAYSLK 129
Query 158 MGGGCGKVCYIDTEATFRPEKICQIAE 184
V Y+DT ++ Q+ +
Sbjct 130 Q-----TVLYVDTTGGLTASRLLQLVQ 151
> cpv:cgd2_4070 hypothetical protein
Length=304
Score = 38.5 bits (88), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 43/86 (50%), Gaps = 14/86 (16%)
Query 99 ELIARRAKVL----KITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQL 154
E+I+ ++L +++TGS +D GGI + EI GE GKTQ C TL +T+ L
Sbjct 23 EIISTSEQMLMDDTRLSTGSNVVDKAFNGGIPKRILFEITGEAGTGKTQWCLTL-ITSVL 81
Query 155 PR---------EMGGGCGKVCYIDTE 171
R E+ G VC + TE
Sbjct 82 LRNLDFSKVMGELNSDIGIVCVLYTE 107
> hsa:5892 RAD51L3, R51H3, RAD51D, TRAD; RAD51-like 3 (S. cerevisiae);
K10871 RAD51-like protein 3
Length=348
Score = 37.4 bits (85), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 5/71 (7%)
Query 114 SPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEAT 173
+ +LD LL G+ T +TEI G GKTQ+C LC+ A + G V Y+D+
Sbjct 106 TSRLDKLLDAGLYTGEVTEIVGGPGSGKTQVC--LCMAANVAH---GLQQNVLYVDSNGG 160
Query 174 FRPEKICQIAE 184
++ Q+ +
Sbjct 161 LTASRLLQLLQ 171
> ath:AT1G79050 DNA repair protein recA
Length=343
Score = 36.6 bits (83), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 46/102 (45%), Gaps = 12/102 (11%)
Query 79 EKIVEAAMKLELCNSFISGGEL---IARRAKVLKITTGSPQLDLLLGGGIETMSITEIFG 135
+K +EAAM ++ +SF G A A V ++G LDL LGGG+ + EI+G
Sbjct 84 QKALEAAMN-DINSSFGKGSVTRLGSAGGALVETFSSGILTLDLALGGGLPKGRVVEIYG 142
Query 136 ENRCGKTQIC-HTLCVTAQLPREMGGGCGKVCYIDTEATFRP 176
GKT + H + +L G +D E F P
Sbjct 143 PESSGKTTLALHAIAEVQKL-------GGNAMLVDAEHAFDP 177
> bbo:BBOV_II001330 18.m06101; hypothetical protein
Length=274
Score = 36.2 bits (82), Expect = 0.087, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 0/75 (0%)
Query 110 ITTGSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYID 169
I+ G ++D LG + +TEI+GE+ GKTQ+ TL + + + Y
Sbjct 13 ISLGITEIDDALGDCLLLGMLTEIYGESGSGKTQVALTLVAEELVRMQEADSNDVMLYFQ 72
Query 170 TEATFRPEKICQIAE 184
T F ++ C I E
Sbjct 73 TSRAFPMQRFCDIIE 87
> cpv:cgd2_3880 hypothetical protein
Length=497
Score = 35.0 bits (79), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 139 CGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEATFRPE 177
GK+ + L + LP E+GG KV YIDT++ F E
Sbjct 106 SGKSLLIMHLIAISILPEEIGGHDQKVYYIDTDSGFSIE 144
> bbo:BBOV_I000620 16.m00943; hypothetical protein
Length=327
Score = 35.0 bits (79), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 45/98 (45%), Gaps = 10/98 (10%)
Query 21 LNEGNDEHFICIDKLQSVGINAADIAKLRGSGYCTVLSVIQTTKKDLALVKGLSEAKVEK 80
L+ GN +H I +LQ IN DIA L + T+ SV V +S +
Sbjct 171 LSHGNRQHCKRIYELQEWCINNVDIASLSAESFATLSSV----------VTYISNPSLSS 220
Query 81 IVEAAMKLELCNSFISGGELIARRAKVLKITTGSPQLD 118
IV A ++ + + + + + K+LK+ T P++D
Sbjct 221 IVPAMIQHFVESYPVEHPQFSSTALKLLKVCTFHPKVD 258
> tpv:TP04_0453 hypothetical protein
Length=286
Score = 34.7 bits (78), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 0/82 (0%)
Query 113 GSPQLDLLLGGGIETMSITEIFGENRCGKTQICHTLCVTAQLPREMGGGCGKVCYIDTEA 172
G ++D L GG+ + EI+G + GKTQ +L + + V YI T
Sbjct 31 GVKEIDQALNGGLLLGKVCEIYGPSGSGKTQFALSLTSEVLINNLIHSKDYVVLYIYTNG 90
Query 173 TFRPEKICQIAERFGLDGAGCL 194
TF E++ +I D G +
Sbjct 91 TFPIERLNEILRSKYEDAKGLI 112
> ath:AT2G19490 recA family protein; K03553 recombination protein
RecA
Length=430
Score = 34.3 bits (77), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 42/88 (47%), Gaps = 11/88 (12%)
Query 89 ELCNSFISGGELIARRA----KVLKITTGSPQLDLLLG-GGIETMSITEIFGENRCGKTQ 143
++ +SF G + RA V +TGS LD+ LG GG+ + EI+G GKT
Sbjct 68 QITSSFGKGSIMYLGRAVSPRNVPVFSTGSFALDVALGVGGLPKGRVVEIYGPEASGKTT 127
Query 144 ICHTLCVTAQLPREMGGGCGKVCYIDTE 171
+ + AQ + GG C ++D E
Sbjct 128 LALHVIAEAQ---KQGGTC---VFVDAE 149
> cpv:cgd4_2050 hypothetical protein
Length=133
Score = 34.3 bits (77), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 21/90 (23%), Positives = 39/90 (43%), Gaps = 12/90 (13%)
Query 140 GKTQICHTLCVTAQLPREMGGGCGKVCYI-DTEATFRPEKICQIAERF-----------G 187
GKT +C L + Q+P+ +GG YI D+E F ++ +I++
Sbjct 21 GKTLLCKILALNIQIPKSIGGPGLNAIYIGDSEGGFSDNRLREISKSTLNYINAKKKTED 80
Query 188 LDGAGCLDNIMYARAYTTEHMHQLFTVAAA 217
+ + NI Y R + E + + T+ +
Sbjct 81 MTCENLIKNIKYIRIFDLEELINVLTLLPS 110
Lambda K H
0.319 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7395169300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40