bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4563_orf3
Length=85
Score E
Sequences producing significant alignments: (Bits) Value
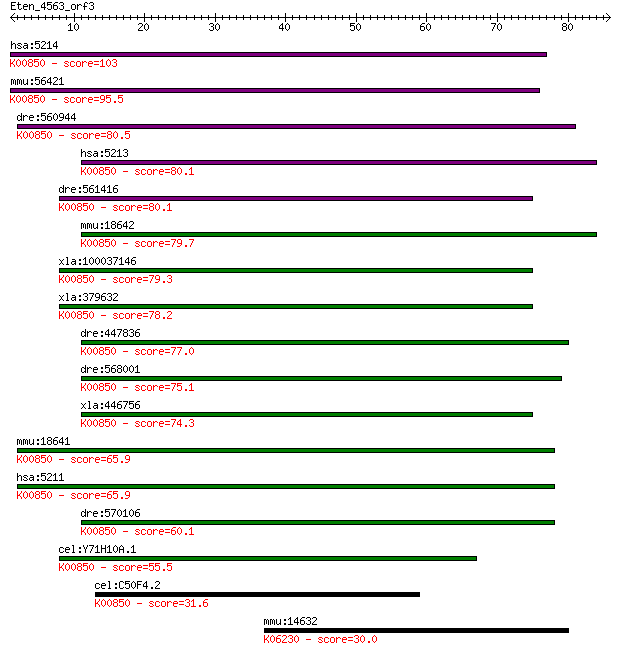
hsa:5214 PFKP, FLJ40226, PFK-C, PFKF; phosphofructokinase, pla... 103 1e-22
mmu:56421 Pfkp, 1200015H23Rik, 9330125N24Rik, PFK-C; phosphofr... 95.5 3e-20
dre:560944 6-phosphofructokinase type C-like; K00850 6-phospho... 80.5 1e-15
hsa:5213 PFKM, GSD7, MGC8699, PFK-1, PFK1, PFKA, PFKX; phospho... 80.1 1e-15
dre:561416 6-phosphofructokinase type C-like; K00850 6-phospho... 80.1 2e-15
mmu:18642 Pfkm, AI131669, PFK-A, PFK-M, Pfk-4, Pfk4, Pfka, Pfk... 79.7 2e-15
xla:100037146 pfkp, pfk, pfk-c, pfkf; phosphofructokinase, pla... 79.3 3e-15
xla:379632 hypothetical protein MGC68847; K00850 6-phosphofruc... 78.2 6e-15
dre:447836 pfkma, pfkm, zgc:92344; phosphofructokinase, muscle... 77.0 1e-14
dre:568001 pfkmb, MGC162168, im:7137110, wu:fc58g11; phosphofr... 75.1 5e-14
xla:446756 pfkm, MGC79063, pfk; phosphofructokinase, muscle (E... 74.3 9e-14
mmu:18641 Pfkl, AA407869; phosphofructokinase, liver, B-type (... 65.9 3e-11
hsa:5211 PFKL, DKFZp686G1648, DKFZp686L2097, FLJ30173, FLJ4090... 65.9 3e-11
dre:570106 6-phosphofructokinase, liver type-like; K00850 6-ph... 60.1 1e-09
cel:Y71H10A.1 hypothetical protein; K00850 6-phosphofructokina... 55.5 4e-08
cel:C50F4.2 hypothetical protein; K00850 6-phosphofructokinase... 31.6 0.58
mmu:14632 Gli1, AV235269, Zfp-5, Zfp5; GLI-Kruppel family memb... 30.0 2.1
> hsa:5214 PFKP, FLJ40226, PFK-C, PFKF; phosphofructokinase, platelet
(EC:2.7.1.11); K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=784
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 49/76 (64%), Positives = 59/76 (77%), Gaps = 0/76 (0%)
Query 1 GKGKKFLSEASGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYR 60
G+GKKF ++ S CVLGISKR ++FQP ELKK+T+ E RIPKEQW LKL PLMK LAKY+
Sbjct 706 GRGKKFTTDDSICVLGISKRNVIFQPVAELKKQTDFEHRIPKEQWWLKLRPLMKILAKYK 765
Query 61 VSFGGSDASQREPVQP 76
S+ SD+ Q E VQP
Sbjct 766 ASYDVSDSGQLEHVQP 781
> mmu:56421 Pfkp, 1200015H23Rik, 9330125N24Rik, PFK-C; phosphofructokinase,
platelet (EC:2.7.1.11); K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=784
Score = 95.5 bits (236), Expect = 3e-20, Method: Composition-based stats.
Identities = 47/75 (62%), Positives = 55/75 (73%), Gaps = 0/75 (0%)
Query 1 GKGKKFLSEASGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYR 60
G GKKF+S+ S CVLGI KR LLFQP ELKK T+ E RIPKEQW LKL P+MK LAKY
Sbjct 705 GTGKKFVSDDSICVLGICKRDLLFQPVAELKKVTDFEHRIPKEQWWLKLRPIMKILAKYE 764
Query 61 VSFGGSDASQREPVQ 75
S+ SD+ + E +Q
Sbjct 765 ASYDMSDSGKLESLQ 779
> dre:560944 6-phosphofructokinase type C-like; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=785
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 41/83 (49%), Positives = 56/83 (67%), Gaps = 4/83 (4%)
Query 2 KGKKFL-SEASGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYR 60
+G+ F +E S C+LG+ +R ++FQP +LK ET+ RIPKEQW LKL PLMK LAKY+
Sbjct 698 RGRVFANTEDSACLLGMRRRAMVFQPVVQLKDETDFVHRIPKEQWWLKLRPLMKILAKYK 757
Query 61 VSFGGSDASQREPV---QPRGPE 80
S+ SD+ Q E V +P+ E
Sbjct 758 TSYDVSDSGQLEHVVVNRPKASE 780
> hsa:5213 PFKM, GSD7, MGC8699, PFK-1, PFK1, PFKA, PFKX; phosphofructokinase,
muscle (EC:2.7.1.11); K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=780
Score = 80.1 bits (196), Expect = 1e-15, Method: Composition-based stats.
Identities = 38/73 (52%), Positives = 48/73 (65%), Gaps = 0/73 (0%)
Query 11 SGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSDASQ 70
SGCVLG+ KR L+FQP ELK +T+ E RIPKEQW LKL P++K LAKY + SD +
Sbjct 707 SGCVLGMRKRALVFQPVAELKDQTDFEHRIPKEQWWLKLRPILKILAKYEIDLDTSDHAH 766
Query 71 REPVQPRGPEEPA 83
E + + E A
Sbjct 767 LEHITRKRSGEAA 779
> dre:561416 6-phosphofructokinase type C-like; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=782
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 37/67 (55%), Positives = 48/67 (71%), Gaps = 0/67 (0%)
Query 8 SEASGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSD 67
+E S C+LG+ +R LLFQP +LK ET+ RIPKEQW L+L PLMK LAKY+ S+ SD
Sbjct 705 TEDSACLLGMRRRALLFQPVVQLKDETDFVHRIPKEQWWLRLRPLMKILAKYKTSYDVSD 764
Query 68 ASQREPV 74
+ Q E +
Sbjct 765 SGQLEHI 771
> mmu:18642 Pfkm, AI131669, PFK-A, PFK-M, Pfk-4, Pfk4, Pfka, Pfkx;
phosphofructokinase, muscle (EC:2.7.1.11); K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=780
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 38/73 (52%), Positives = 48/73 (65%), Gaps = 0/73 (0%)
Query 11 SGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSDASQ 70
SGCVLG+ KR L+FQP ELK +T+ E RIPKEQW LKL P++K LAKY + SD +
Sbjct 707 SGCVLGMRKRALVFQPVTELKDQTDFEHRIPKEQWWLKLRPILKILAKYEIDLDTSDHAH 766
Query 71 REPVQPRGPEEPA 83
E + + E A
Sbjct 767 LEHISRKRSGEAA 779
> xla:100037146 pfkp, pfk, pfk-c, pfkf; phosphofructokinase, platelet
(EC:2.7.1.11); K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=786
Score = 79.3 bits (194), Expect = 3e-15, Method: Composition-based stats.
Identities = 37/67 (55%), Positives = 49/67 (73%), Gaps = 0/67 (0%)
Query 8 SEASGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSD 67
++ S C+LG+ +R LLFQP +LK ET+ + RIPKEQW LK+ PLMK LAKY+ S+ SD
Sbjct 708 TDDSVCLLGMRRRNLLFQPVTQLKDETDFKHRIPKEQWWLKMRPLMKILAKYKTSYDISD 767
Query 68 ASQREPV 74
A + E V
Sbjct 768 AGKLEHV 774
> xla:379632 hypothetical protein MGC68847; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=784
Score = 78.2 bits (191), Expect = 6e-15, Method: Composition-based stats.
Identities = 36/67 (53%), Positives = 49/67 (73%), Gaps = 0/67 (0%)
Query 8 SEASGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSD 67
++ S C+LG+ +R LLFQP +LK ET+ + RIP+EQW LK+ PLMK LAKY+ S+ SD
Sbjct 706 TDDSVCLLGMRRRNLLFQPVTQLKDETDFKHRIPREQWWLKMRPLMKILAKYKTSYDISD 765
Query 68 ASQREPV 74
A + E V
Sbjct 766 AGKLEHV 772
> dre:447836 pfkma, pfkm, zgc:92344; phosphofructokinase, muscle
a (EC:2.7.1.11); K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=784
Score = 77.0 bits (188), Expect = 1e-14, Method: Composition-based stats.
Identities = 36/69 (52%), Positives = 47/69 (68%), Gaps = 0/69 (0%)
Query 11 SGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSDASQ 70
S CVLG+ KR L+ QP ELK+ET+ E RIPK QW LKL P+MK LAKY+++ S+ +
Sbjct 708 SACVLGMRKRGLVLQPLAELKEETDFEHRIPKSQWWLKLRPIMKILAKYKINLDTSEHAL 767
Query 71 REPVQPRGP 79
E V + P
Sbjct 768 MEHVVSKKP 776
> dre:568001 pfkmb, MGC162168, im:7137110, wu:fc58g11; phosphofructokinase,
muscle b; K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=776
Score = 75.1 bits (183), Expect = 5e-14, Method: Composition-based stats.
Identities = 35/69 (50%), Positives = 49/69 (71%), Gaps = 1/69 (1%)
Query 11 SGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSDASQ 70
S CVLG+ KR +LFQP +LK++T+ E RIPK +W LKL P++K LAKY++S S+ +
Sbjct 706 SACVLGMRKRAMLFQPLSDLKEDTDFEHRIPKTEWWLKLRPILKILAKYKISLDTSETAT 765
Query 71 REPV-QPRG 78
E V + RG
Sbjct 766 MEHVIKKRG 774
> xla:446756 pfkm, MGC79063, pfk; phosphofructokinase, muscle
(EC:2.7.1.11); K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=808
Score = 74.3 bits (181), Expect = 9e-14, Method: Composition-based stats.
Identities = 32/64 (50%), Positives = 47/64 (73%), Gaps = 0/64 (0%)
Query 11 SGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSDASQ 70
S C+LG+ KR+L+FQP ELK++T+ R+PKEQW LKL P++K LAKY+++ SD +
Sbjct 735 SACLLGMRKRSLVFQPLVELKEQTDFAHRLPKEQWWLKLRPILKILAKYKINLDTSDHAH 794
Query 71 REPV 74
E +
Sbjct 795 MEHI 798
> mmu:18641 Pfkl, AA407869; phosphofructokinase, liver, B-type
(EC:2.7.1.11); K00850 6-phosphofructokinase [EC:2.7.1.11]
Length=780
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 33/77 (42%), Positives = 47/77 (61%), Gaps = 1/77 (1%)
Query 2 KGKKFLSEA-SGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYR 60
KG+ F + S CV+G+ K+ + F P ELKKET+ E R+P+EQW L L ++K LA YR
Sbjct 696 KGRVFANAPDSACVIGLRKKVVAFSPVTELKKETDFEHRMPREQWWLNLRLMLKMLAHYR 755
Query 61 VSFGGSDASQREPVQPR 77
+S + + E V R
Sbjct 756 ISMADYVSGELEHVTRR 772
> hsa:5211 PFKL, DKFZp686G1648, DKFZp686L2097, FLJ30173, FLJ40909,
PFK-B; phosphofructokinase, liver (EC:2.7.1.11); K00850
6-phosphofructokinase [EC:2.7.1.11]
Length=780
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 32/77 (41%), Positives = 48/77 (62%), Gaps = 1/77 (1%)
Query 2 KGKKFLSEA-SGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYR 60
KG+ F + S CV+G+ K+ + F P ELKK+T+ E R+P+EQW L L ++K LA+YR
Sbjct 696 KGRVFANAPDSACVIGLKKKAVAFSPVTELKKDTDFEHRMPREQWWLSLRLMLKMLAQYR 755
Query 61 VSFGGSDASQREPVQPR 77
+S + + E V R
Sbjct 756 ISMAAYVSGELEHVTRR 772
> dre:570106 6-phosphofructokinase, liver type-like; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=780
Score = 60.1 bits (144), Expect = 1e-09, Method: Composition-based stats.
Identities = 28/67 (41%), Positives = 41/67 (61%), Gaps = 0/67 (0%)
Query 11 SGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGSDASQ 70
+ CV+G+ K+ + F P ELK++T+ E R+PK QW L L ++K LAKY+ SF +
Sbjct 706 TACVVGLYKKAMTFSPVTELKEQTDFEHRMPKTQWWLNLRLMLKMLAKYQTSFNEYVTGE 765
Query 71 REPVQPR 77
E V R
Sbjct 766 IEHVTRR 772
> cel:Y71H10A.1 hypothetical protein; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=814
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 24/59 (40%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 8 SEASGCVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAKYRVSFGGS 66
S + +LG+ R ++F P +LKKET+ E R+P EQW + L PL++ LA++R + S
Sbjct 740 SAHTATLLGLKGRKVVFTPVQDLKKETDFEHRLPSEQWWMALRPLLRVLARHRSTVESS 798
> cel:C50F4.2 hypothetical protein; K00850 6-phosphofructokinase
[EC:2.7.1.11]
Length=756
Score = 31.6 bits (70), Expect = 0.58, Method: Composition-based stats.
Identities = 15/46 (32%), Positives = 24/46 (52%), Gaps = 0/46 (0%)
Query 13 CVLGISKRTLLFQPGGELKKETNLEPRIPKEQWGLKLGPLMKFLAK 58
CV+G+ +L + P L K+ E +P W L L PL++ + K
Sbjct 702 CVIGLRGSSLRYVPVQGLGKKVCFEHGVPHNMWWLDLHPLVEAMTK 747
> mmu:14632 Gli1, AV235269, Zfp-5, Zfp5; GLI-Kruppel family member
GLI1; K06230 zinc finger protein GLI
Length=1111
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 37 EPRIPKEQWGLKLGPLMKFLAKYRVSFGGSDASQREPVQPRGP 79
EP + G L P M F + + +GG + + EP + RGP
Sbjct 707 EPEVGTSVMGNGLNPYMDFSSTDTLGYGGPEGTAAEPYEARGP 749
Lambda K H
0.315 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2035463248
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40