bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4538_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
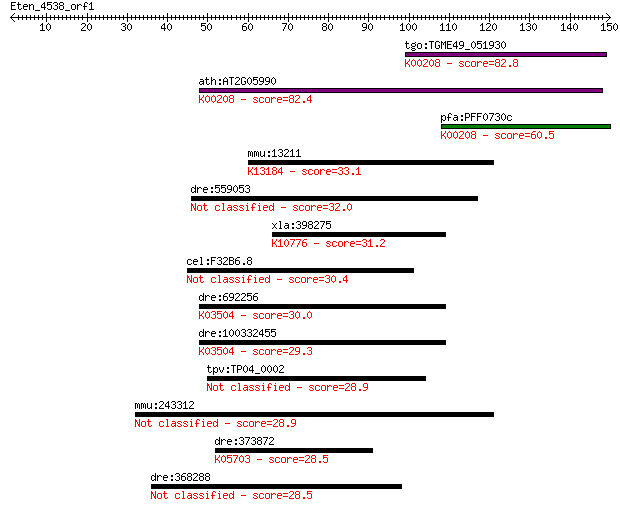
tgo:TGME49_051930 enoyl-acyl carrier reductase (EC:1.3.1.9); K... 82.8 3e-16
ath:AT2G05990 MOD1; MOD1 (MOSAIC DEATH 1); enoyl-[acyl-carrier... 82.4 4e-16
pfa:PFF0730c PfENR; enoyl-acyl carrier reductase (EC:1.3.1.9);... 60.5 2e-09
mmu:13211 Dhx9, AI326842, Ddx9, HEL-5, MGC90954, NDHII, RHA, m... 33.1 0.36
dre:559053 casz1, sb:eu580, wu:fk85f10; castor zinc finger 1 32.0
xla:398275 lig3; ligase III, DNA, ATP-dependent (EC:6.5.1.1); ... 31.2 1.3
cel:F32B6.8 tbc-3; TBC (Tre-2/Bub2/Cdc16) domain family member... 30.4 2.4
dre:692256 pold3, zgc:136471; polymerase (DNA-directed), delta... 30.0 2.8
dre:100332455 DNA-directed DNA polymerase delta 3-like; K03504... 29.3 4.4
tpv:TP04_0002 hypothetical protein 28.9 6.2
mmu:243312 Elfn1, A930017N06Rik, AI197272; leucine rich repeat... 28.9 6.9
dre:373872 fyna, c-fyn, fyn, zgc:86720; FYN oncogene related t... 28.5 7.7
dre:368288 rassf7b, sb:cb262, zgc:55456; Ras association (RalG... 28.5 8.2
> tgo:TGME49_051930 enoyl-acyl carrier reductase (EC:1.3.1.9);
K00208 enoyl-[acyl-carrier protein] reductase I [EC:1.3.1.9]
Length=417
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 36/50 (72%), Positives = 43/50 (86%), Gaps = 0/50 (0%)
Query 99 LPVDLRGKTAFVAGVADSNGYGWAICKLLRAAGARVLVGTWPPVYSIFKK 148
P+DLRG+TAFVAGVADS+GYGWAI K L +AGARV +GTWPPV +F+K
Sbjct 105 FPIDLRGQTAFVAGVADSHGYGWAIAKHLASAGARVALGTWPPVLGLFQK 154
> ath:AT2G05990 MOD1; MOD1 (MOSAIC DEATH 1); enoyl-[acyl-carrier-protein]
reductase (NADH)/ enoyl-[acyl-carrier-protein] reductase/
oxidoreductase (EC:1.3.1.10); K00208 enoyl-[acyl-carrier
protein] reductase I [EC:1.3.1.9]
Length=390
Score = 82.4 bits (202), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 45/100 (45%), Positives = 58/100 (58%), Gaps = 19/100 (19%)
Query 48 LRPTAAVPTASQQQQPVQQVAVVPRGVSSSSSSSTAAAAGPTGTAEPLSGPLPVDLRGKT 107
LR +AVPT + P S+ + S ++ P+G LP+DLRGK
Sbjct 56 LRNHSAVPTCKR-----------PFSFSTRAMSESSENKAPSG--------LPIDLRGKR 96
Query 108 AFVAGVADSNGYGWAICKLLRAAGARVLVGTWPPVYSIFK 147
AF+AG+AD NGYGWAI K L AAGA +LVGTW P +IF+
Sbjct 97 AFIAGIADDNGYGWAIAKSLAAAGAEILVGTWVPALNIFE 136
> pfa:PFF0730c PfENR; enoyl-acyl carrier reductase (EC:1.3.1.9);
K00208 enoyl-[acyl-carrier protein] reductase I [EC:1.3.1.9]
Length=432
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 21/42 (50%), Positives = 28/42 (66%), Gaps = 0/42 (0%)
Query 108 AFVAGVADSNGYGWAICKLLRAAGARVLVGTWPPVYSIFKKG 149
F+AG+ D+NGYGW I K L +++ G WPPVY+IF K
Sbjct 100 CFIAGIGDTNGYGWGIAKELSKRNVKIIFGIWPPVYNIFMKN 141
> mmu:13211 Dhx9, AI326842, Ddx9, HEL-5, MGC90954, NDHII, RHA,
mHEL-5; DEAH (Asp-Glu-Ala-His) box polypeptide 9 (EC:3.6.4.13);
K13184 ATP-dependent RNA helicase A [EC:3.6.4.13]
Length=1383
Score = 33.1 bits (74), Expect = 0.36, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query 60 QQQPVQQVAVVPRG-VSSSSSSSTAAAAGPTGTAEPLSGPLPVDLRGKTAFVAGVADSNG 118
+ + V V +VP + S +S STA+AA G P+ GPLP L K +S+G
Sbjct 76 KSEEVPAVGIVPPPPILSDTSDSTASAA--EGLPAPMGGPLPPHLALKAEENNSGVESSG 133
Query 119 YG 120
YG
Sbjct 134 YG 135
> dre:559053 casz1, sb:eu580, wu:fk85f10; castor zinc finger 1
Length=1760
Score = 32.0 bits (71), Expect = 0.71, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 35/71 (49%), Gaps = 2/71 (2%)
Query 46 QRLRPTAAVPTASQQQQPVQQVAVVPRGVSSSSSSSTAAAAGPTGTAEPLSGPLPVDLRG 105
Q ++PTAA+P AS+ + Q +P V + + S +A AG + PL +P+ L
Sbjct 744 QSVKPTAALPPASRISTLLSQ--ALPSNVPMALALSNSALAGASNAFFPLIPRMPMPLPP 801
Query 106 KTAFVAGVADS 116
A + A S
Sbjct 802 SAAGLISAATS 812
> xla:398275 lig3; ligase III, DNA, ATP-dependent (EC:6.5.1.1);
K10776 DNA ligase 3 [EC:6.5.1.1]
Length=988
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 66 QVAVVPRGVSSSSSSSTAAAAGPTGTAEPLSGPLPVDLRGKTA 108
+ A PR + S + + AA G T T P + P P+ G +A
Sbjct 155 KTAATPRKKTPSKGNQSMAAQGTTPTKAPATSPSPLKFSGYSA 197
> cel:F32B6.8 tbc-3; TBC (Tre-2/Bub2/Cdc16) domain family member
(tbc-3)
Length=475
Score = 30.4 bits (67), Expect = 2.4, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 7/56 (12%)
Query 45 QQRLRPTAAVPTASQQQQPVQQVAVVPRGVSSSSSSSTAAAAGPTGTAEPLSGPLP 100
++ + P +A PT + +QQ +Q++A+ + SS SS+ TA P+ LP
Sbjct 64 EEIMPPVSAAPTKNSRQQALQRLAIQKEPLPSSVSSTPQ-------TANPIYPKLP 112
> dre:692256 pold3, zgc:136471; polymerase (DNA-directed), delta
3, accessory subunit; K03504 DNA polymerase delta subunit
3
Length=457
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 4/61 (6%)
Query 48 LRPTAAVPTASQQQQPVQQVAVVPRGVSSSSSSSTAAAAGPTGTAEPLSGPLPVDLRGKT 107
+R AAVP +S + Q Q++A+VP S+ + P T++P + P P + G
Sbjct 134 IRCAAAVPMSSAELQRAQEIALVP---SAEKENRKPGLNEPAATSKPSAKP-PAGIMGMF 189
Query 108 A 108
A
Sbjct 190 A 190
> dre:100332455 DNA-directed DNA polymerase delta 3-like; K03504
DNA polymerase delta subunit 3
Length=457
Score = 29.3 bits (64), Expect = 4.4, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 31/61 (50%), Gaps = 4/61 (6%)
Query 48 LRPTAAVPTASQQQQPVQQVAVVPRGVSSSSSSSTAAAAGPTGTAEPLSGPLPVDLRGKT 107
+R AAVP +S + Q Q++A+VP S+ + P T++P + P P + G
Sbjct 134 IRCAAAVPMSSAELQRAQEMALVP---SAEKENRKPGLNEPAATSKPSAKP-PAGIMGMF 189
Query 108 A 108
A
Sbjct 190 A 190
> tpv:TP04_0002 hypothetical protein
Length=472
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 24/54 (44%), Gaps = 1/54 (1%)
Query 50 PTAAVPTASQQQQPVQQVAVVPRGVSSSSSSSTAAAAGPTGTAEPLSGPLPVDL 103
P ++ +Q QP+ Q P+ + + T P EP+ P+PVD+
Sbjct 197 PQQSIEQPIEQPQPIDQPTEQPQPIDQPAEE-TEPIDQPIEQPEPIDQPMPVDI 249
> mmu:243312 Elfn1, A930017N06Rik, AI197272; leucine rich repeat
and fibronectin type III, extracellular 1
Length=828
Score = 28.9 bits (63), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 42/94 (44%), Gaps = 5/94 (5%)
Query 32 RVGPGPEARGAELQQRLRPTAAVPTASQQQQPVQQVAVVPRG----VSSSSSSSTAAAAG 87
R G E RG EL + P ++V S + V +V + + S S+S A +G
Sbjct 529 RTGEPQERRGCELSRPGEPQSSVAEISTIAKEVDRVNQIINNCIDALKSESTSFQGAKSG 588
Query 88 PTGTAEPLSGPLPVDLRGKTAFVAGV-ADSNGYG 120
AEP L L K +F++ V D+ G+G
Sbjct 589 AVSAAEPQLVLLSEPLASKHSFLSPVYKDAFGHG 622
> dre:373872 fyna, c-fyn, fyn, zgc:86720; FYN oncogene related
to SRC, FGR, YES a; K05703 proto-oncogene tyrosine-protein
kinase Fyn [EC:2.7.10.2]
Length=293
Score = 28.5 bits (62), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 52 AAVPTASQQQQPVQQVAVVPRGVSSSSSSSTAAAAGPTG 90
A+P + PV Q V GV+SSS S T + G TG
Sbjct 45 TAIPNYNNFHAPVSQGVTVFGGVNSSSHSGTLRSRGGTG 83
> dre:368288 rassf7b, sb:cb262, zgc:55456; Ras association (RalGDS/AF-6)
domain family (N-terminal) member 7b
Length=427
Score = 28.5 bits (62), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 28/63 (44%), Gaps = 1/63 (1%)
Query 36 GPEARGAELQQRLRPTAAVPTASQQQQPVQQVAVV-PRGVSSSSSSSTAAAAGPTGTAEP 94
E R LQQ ++ T P S QQ A++ P G S S+S+ P TA+
Sbjct 339 NKELRQCNLQQFIQQTGVTPAQSSQQTEEMDFALLKPDGQSDEDSTSSILEFNPRTTAKQ 398
Query 95 LSG 97
+ G
Sbjct 399 ILG 401
Lambda K H
0.317 0.132 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3068761412
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40