bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4527_orf3
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
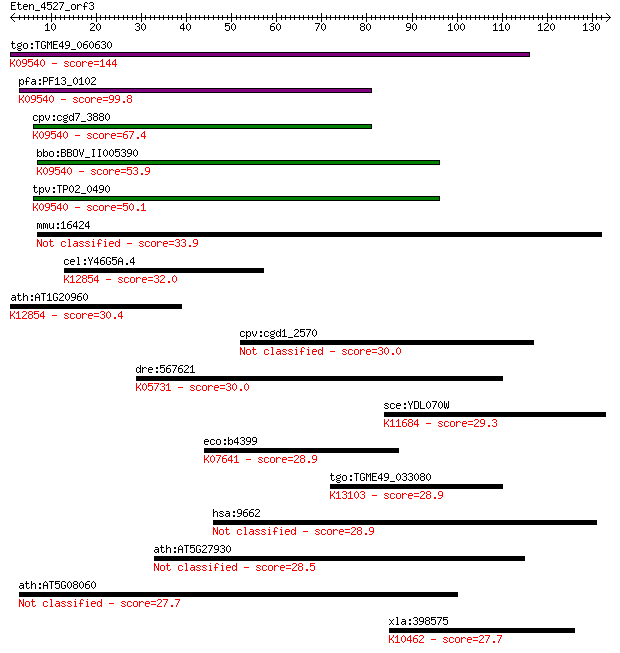
tgo:TGME49_060630 DnaJ domain-containing protein ; K09540 tran... 144 5e-35
pfa:PF13_0102 DnaJ/SEC63 protein, putative; K09540 translocati... 99.8 2e-21
cpv:cgd7_3880 DNAJ domain protein sec63 ortholog, 4 transmembr... 67.4 1e-11
bbo:BBOV_II005390 18.m06448; DnaJ domain containing protein; K... 53.9 1e-07
tpv:TP02_0490 hypothetical protein; K09540 translocation prote... 50.1 2e-06
mmu:16424 Itih1, Intin1, Itih-1; inter-alpha trypsin inhibitor... 33.9 0.13
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 32.0 0.56
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 30.4 1.6
cpv:cgd1_2570 alpha beta hydrolase 30.0 2.0
dre:567621 tiam1; T-cell lymphoma invasion and metastasis 1; K... 30.0 2.1
sce:YDL070W BDF2; Bdf2p; K11684 bromodomain-containing factor 1 29.3
eco:b4399 creC, ECK4391, JW4362, phoM; sensory histidine kinas... 28.9 4.1
tgo:TGME49_033080 tuftelin interacting protein 11, putative (E... 28.9 4.3
hsa:9662 CEP135, CEP4, KIAA0635; centrosomal protein 135kDa 28.9 5.0
ath:AT5G27930 protein phosphatase 2C, putative / PP2C, putative 28.5 5.6
ath:AT5G08060 hypothetical protein 27.7 8.9
xla:398575 klhl25, Xencr-2, encr-2; kelch-like 25; K10462 kelc... 27.7 8.9
> tgo:TGME49_060630 DnaJ domain-containing protein ; K09540 translocation
protein SEC63
Length=675
Score = 144 bits (364), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 70/115 (60%), Positives = 88/115 (76%), Gaps = 1/115 (0%)
Query 1 KSRNLERVVKESLQFRVGRPGKNVMTVVALCDSYTGVDATIDVEFRAYTPEEKPREVFVH 60
+S++ ERVV+E +QFRV R GK +TV+A+CDSY G D T+++EF+AY PEEKPR V++H
Sbjct 525 RSKSAERVVEERIQFRVNRVGKQSVTVLAICDSYAGCDCTMELEFKAYHPEEKPRPVWIH 584
Query 61 PEDVKLDEEPTLFQQMLGDMCVNSSDEEEEFDLDEAPKVVRKKVATENDGVATAE 115
PED++LDEEPTLFQQMLG+M SSDEEE FDLDE +V KK E A AE
Sbjct 585 PEDLRLDEEPTLFQQMLGEM-YTSSDEEESFDLDEGLRVAVKKKTQEAGEAANAE 638
> pfa:PF13_0102 DnaJ/SEC63 protein, putative; K09540 translocation
protein SEC63
Length=651
Score = 99.8 bits (247), Expect = 2e-21, Method: Composition-based stats.
Identities = 43/78 (55%), Positives = 59/78 (75%), Gaps = 0/78 (0%)
Query 3 RNLERVVKESLQFRVGRPGKNVMTVVALCDSYTGVDATIDVEFRAYTPEEKPREVFVHPE 62
+N E++++E LQF V + G ++V ALCDSY G D +D+ F+AY+ E RE+FVHPE
Sbjct 521 KNCEKIIEEKLQFLVDKVGNLSVSVFALCDSYFGCDQKVDIPFKAYSKTEIKREIFVHPE 580
Query 63 DVKLDEEPTLFQQMLGDM 80
D++LD EPTLFQQMLGD+
Sbjct 581 DIELDNEPTLFQQMLGDI 598
> cpv:cgd7_3880 DNAJ domain protein sec63 ortholog, 4 transmembrane
domains ; K09540 translocation protein SEC63
Length=627
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 30/75 (40%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 6 ERVVKESLQFRVGRPGKNVMTVVALCDSYTGVDATIDVEFRAYTPEEKPREVFVHPEDVK 65
E++V +QF + PG +++ + DSY G+D ++V F A T +E R+++VHPED
Sbjct 529 EKIVDVKIQFLIETPGSIDISLHLINDSYEGLDQVVNVSFVAKTIKEGIRQIYVHPEDEA 588
Query 66 LDEEPTLFQQMLGDM 80
LD EPTLFQ ++ +
Sbjct 589 LDNEPTLFQHIMNQL 603
> bbo:BBOV_II005390 18.m06448; DnaJ domain containing protein;
K09540 translocation protein SEC63
Length=618
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/89 (31%), Positives = 46/89 (51%), Gaps = 1/89 (1%)
Query 7 RVVKESLQFRVGRPGKNVMTVVALCDSYTGVDATIDVEFRAYTPEEKPREVFVHPEDVKL 66
RV+ E + G + + V A+ DSY G D + VEF + + +HP+D+ L
Sbjct 531 RVITEKISVLAENVGMHSVLVTAMSDSYFGCDKSTRVEFMVNMANDA-LDFKIHPDDIAL 589
Query 67 DEEPTLFQQMLGDMCVNSSDEEEEFDLDE 95
D EP+ +M+GD+ E+EE +D+
Sbjct 590 DNEPSAIGKMIGDLLRQEDSEDEEEVIDD 618
> tpv:TP02_0490 hypothetical protein; K09540 translocation protein
SEC63
Length=659
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 29/92 (31%), Positives = 49/92 (53%), Gaps = 2/92 (2%)
Query 6 ERVVKESLQFRVGRPGKNVMTVVALCDSYTGVDATIDVEF--RAYTPEEKPREVFVHPED 63
E V E +Q + G+N + V A+ DSY G + ++ +F ++K +HPED
Sbjct 568 EAEVVEKMQILAEKVGQNTICVTAVNDSYFGAEFSVLKKFYVNPLNLQKKYESFKIHPED 627
Query 64 VKLDEEPTLFQQMLGDMCVNSSDEEEEFDLDE 95
+KLDE+ +M+G++ S +EEE + E
Sbjct 628 MKLDEDVNPITKMIGELLTAESSDEEEVEYIE 659
> mmu:16424 Itih1, Intin1, Itih-1; inter-alpha trypsin inhibitor,
heavy chain 1
Length=907
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 39/144 (27%), Positives = 58/144 (40%), Gaps = 27/144 (18%)
Query 7 RVVKESLQFRVGRPGKNVMTVVALCDSYTGVDATIDVEFRAYTPEEK--PREVFV----- 59
+V+K SL + P + +T+ L D G++ TID P E PR FV
Sbjct 589 QVLKMSLDYHFVTPLTS-LTIRGLTDE-DGLEPTIDKPLEDSQPLEMVGPRRTFVLSAIQ 646
Query 60 -----HPEDVKL-------DEEPTLFQQMLGDMCVNSSDEEEEFDLDEAPKVVRKKVATE 107
HP D KL D +P + V S ++ F+++E P V+ V
Sbjct 647 PSPTAHPIDSKLPLRVTGVDTDPHFI------IYVPSKEDSLCFNINEEPGVILNLVQDP 700
Query 108 NDGVATAEQADSNKAGESGERPTT 131
+ G Q NKA G+ +T
Sbjct 701 DTGFTVNGQLIGNKASSPGQHEST 724
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 32.0 bits (71), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 20/44 (45%), Gaps = 0/44 (0%)
Query 13 LQFRVGRPGKNVMTVVALCDSYTGVDATIDVEFRAYTPEEKPRE 56
L F RPG + + + DSY G D DV F+ P R+
Sbjct 2094 LDFAAPRPGHHKFKLFFISDSYLGADQEFDVAFKVEEPGRSNRK 2137
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 1 KSRNLERVVKESLQFRV-GRPGKNVMTVVALCDSYTGVD 38
K +L+R VK L F PG+ T+ +CDSY G D
Sbjct 2113 KRVSLQRKVKVKLDFTAPSEPGEKSYTLYFMCDSYLGCD 2151
> cpv:cgd1_2570 alpha beta hydrolase
Length=1448
Score = 30.0 bits (66), Expect = 2.0, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Query 52 EKPREVFVHPEDVKLDEEPTLFQQMLGDMCVNSSDEEEEFDLDEAPKVVRKKVATENDGV 111
EK E+ D L+ P +F M D +N S + +EF+ +E P+V K+ +D +
Sbjct 941 EKTNELIHSTSDNVLETIP-IFGNMGFDQ-INQSADTDEFNYEEIPEVDSNKIEKLHDEI 998
Query 112 ATAEQ 116
+ EQ
Sbjct 999 ESPEQ 1003
> dre:567621 tiam1; T-cell lymphoma invasion and metastasis 1;
K05731 T-cell lymphoma invasion and metastasis
Length=1684
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 41/93 (44%), Gaps = 15/93 (16%)
Query 29 ALCDSYTGV-----DATIDVEFRAYTPEEKPR-------EVFVHPEDVKLDEEPTLFQQM 76
A C S+T V A D EF+A+ E PR E ++ ++ + P L +++
Sbjct 1226 AFCASHTKVPKVLTKAKTDPEFKAFLAERNPRQQHSSTLESYLIKPIQRVLKYPLLLREL 1285
Query 77 LGDMCVNSSDEEEEFDLDEAPKVVRKKVATEND 109
+ D EE + LD A K + K + N+
Sbjct 1286 YS---LTDPDSEEHYHLDVAMKAMNKVASHINE 1315
> sce:YDL070W BDF2; Bdf2p; K11684 bromodomain-containing factor
1
Length=638
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 5/49 (10%)
Query 84 SSDEEEEFDLDEAPKVVRKKVATENDGVATAEQADSNKAGESGERPTTT 132
S + ++ D+D +P V+R+ V+T ND + + N+ G SG RP T
Sbjct 253 SRNRKKNEDMD-SPLVIRRSVSTTNDNIGES----GNREGVSGGRPKRT 296
> eco:b4399 creC, ECK4391, JW4362, phoM; sensory histidine kinase
in two-component regulatory system with CreB or PhoB, regulator
of the CreBC regulon; K07641 two-component system, OmpR
family, sensor histidine kinase CreC [EC:2.7.13.3]
Length=474
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 44 EFRAYTPEEKPREVFVHPEDVKLDEEPTLFQQMLGDMCVNSSD 86
E R EK + V P +V + EP L +Q LG++ N+ D
Sbjct 339 EARTVQLAEKKITLHVTPTEVNVAAEPALLEQALGNLLDNAID 381
> tgo:TGME49_033080 tuftelin interacting protein 11, putative
(EC:3.4.21.72); K13103 tuftelin-interacting protein 11
Length=1415
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 24/38 (63%), Gaps = 1/38 (2%)
Query 72 LFQQMLGDMCVNSSDEEEEFDLDEAPKVVRKKVATEND 109
+ +++LGD SS+EEE+ DLDE + K A END
Sbjct 119 VLKEVLGD-AYESSEEEEKADLDEEKEKASAKSAEEND 155
> hsa:9662 CEP135, CEP4, KIAA0635; centrosomal protein 135kDa
Length=1140
Score = 28.9 bits (63), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 40/85 (47%), Gaps = 0/85 (0%)
Query 46 RAYTPEEKPREVFVHPEDVKLDEEPTLFQQMLGDMCVNSSDEEEEFDLDEAPKVVRKKVA 105
R +E RE H E+V L + L + + CVN E E+++L ++++ +
Sbjct 580 RIMAEKEALREKLEHIEEVSLFGKSELEKTIEHLTCVNHQLESEKYELKSKVLIMKETIE 639
Query 106 TENDGVATAEQADSNKAGESGERPT 130
+ + + Q S+ AG+S + T
Sbjct 640 SLENKLKVQAQKFSHVAGDSSHQKT 664
> ath:AT5G27930 protein phosphatase 2C, putative / PP2C, putative
Length=373
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 22/96 (22%), Positives = 40/96 (41%), Gaps = 22/96 (22%)
Query 33 SYTGVDATIDVEFRAYTPEEKPREVFVHPEDVKLDEEPTL--------------FQQMLG 78
S V T+D F+ P+EK R + LD+EP + + G
Sbjct 210 SLVAVQLTLD--FKPNLPQEKERIIGCKGRVFCLDDEPGVHRVWQPDAETPGLAMSRAFG 267
Query 79 DMCVNSSDEEEEFDLDEAPKVVRKKVATENDGVATA 114
D C+ +E+ L P+V ++ ++T++ + A
Sbjct 268 DYCI------KEYGLVSVPEVTQRHISTKDHFIILA 297
> ath:AT5G08060 hypothetical protein
Length=131
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 40/101 (39%), Gaps = 6/101 (5%)
Query 3 RNLERVVKESLQFRVGRPGKNVMTVVALCDSY-TGVDATIDVEFRAYTPEEKPREVF--- 58
+NL R +K+ Q + V Y ATID E A P P V+
Sbjct 14 QNLRRYIKKPWQITGPCAHPEYLEAVPKATEYRLRCPATIDEE--AIVPSSDPETVYNIV 71
Query 59 VHPEDVKLDEEPTLFQQMLGDMCVNSSDEEEEFDLDEAPKV 99
H D + + P + D V +E++ FD+ + PKV
Sbjct 72 YHGRDQRRNRPPIRRYVLTKDNVVQMMNEKKSFDVSDFPKV 112
> xla:398575 klhl25, Xencr-2, encr-2; kelch-like 25; K10462 kelch-like
protein 25/37 (ectoderm-neural cortex protein)
Length=589
Score = 27.7 bits (60), Expect = 8.9, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query 85 SDEEEEFDLDEAPKVVRKKVATENDGVATAEQADSNKAGES 125
+DE + +DEA V KK +NDGV T+ A KAG +
Sbjct 258 ADERSKLIMDEA--VQCKKKILQNDGVVTSLCAKPRKAGHT 296
Lambda K H
0.309 0.128 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40