bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4526_orf2
Length=239
Score E
Sequences producing significant alignments: (Bits) Value
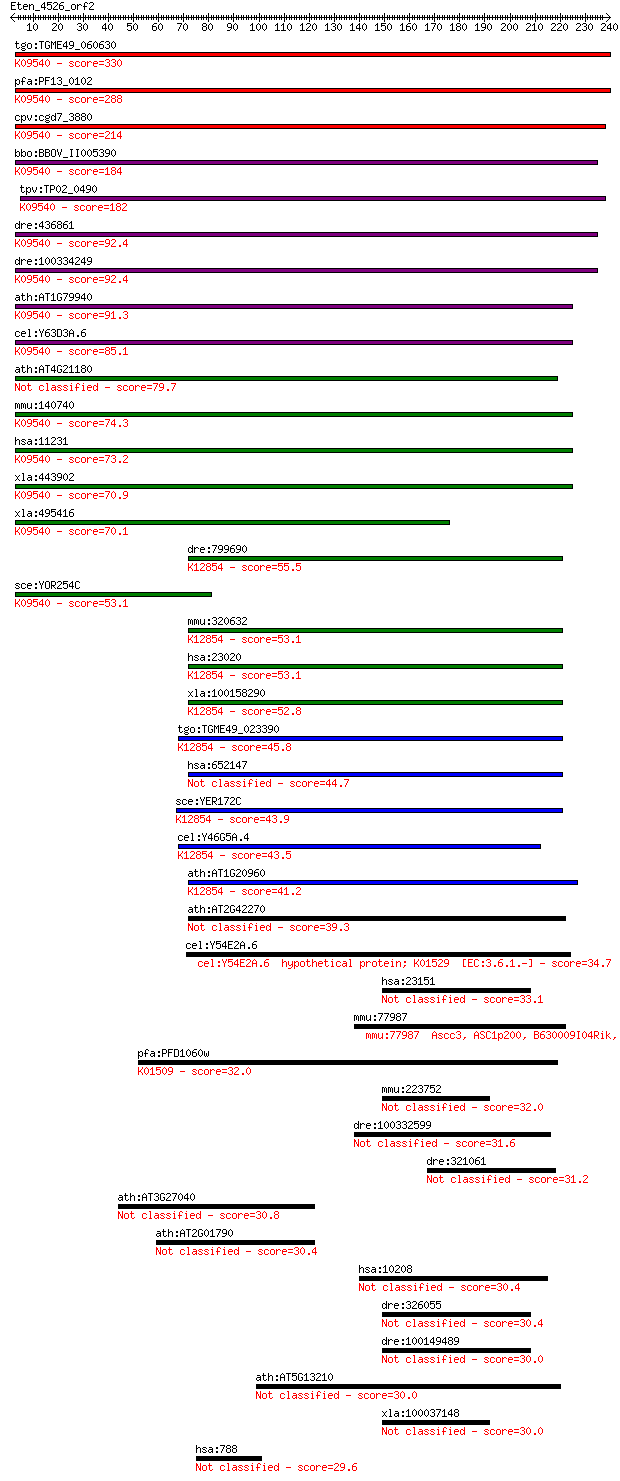
tgo:TGME49_060630 DnaJ domain-containing protein ; K09540 tran... 330 3e-90
pfa:PF13_0102 DnaJ/SEC63 protein, putative; K09540 translocati... 288 9e-78
cpv:cgd7_3880 DNAJ domain protein sec63 ortholog, 4 transmembr... 214 2e-55
bbo:BBOV_II005390 18.m06448; DnaJ domain containing protein; K... 184 2e-46
tpv:TP02_0490 hypothetical protein; K09540 translocation prote... 182 8e-46
dre:436861 sec63, zgc:92718; SEC63-like (S. cerevisiae); K0954... 92.4 1e-18
dre:100334249 SEC63-like protein-like; K09540 translocation pr... 92.4 1e-18
ath:AT1G79940 ATERDJ2A; heat shock protein binding / unfolded ... 91.3 3e-18
cel:Y63D3A.6 dnj-29; DNaJ domain (prokaryotic heat shock prote... 85.1 2e-16
ath:AT4G21180 ATERDJ2B; heat shock protein binding / unfolded ... 79.7 9e-15
mmu:140740 Sec63, 5730478J10Rik, AI649014, AW319215; SEC63-lik... 74.3 4e-13
hsa:11231 SEC63, ERdj2, PRO2507, SEC63L; SEC63 homolog (S. cer... 73.2 7e-13
xla:443902 MGC80164 protein; K09540 translocation protein SEC63 70.9 4e-12
xla:495416 sec63, MGC132056; SEC63 homolog; K09540 translocati... 70.1 7e-12
dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-... 55.5 2e-07
sce:YOR254C SEC63, PTL1; Essential subunit of Sec63 complex (S... 53.1 7e-07
mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,... 53.1 8e-07
hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-... 53.1 8e-07
xla:100158290 snrnp200, ascc3l1; small nuclear ribonucleoprote... 52.8 1e-06
tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helica... 45.8 2e-04
hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase... 44.7 3e-04
sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.... 43.9 5e-04
cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing he... 43.5 6e-04
ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding / A... 41.2 0.003
ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, put... 39.3 0.013
cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-] 34.7 0.32
hsa:23151 GRAMD4, DIP, KIAA0767, MGC149847, MGC90497, dA59H18.... 33.1 0.94
mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07... 32.7 1.1
pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific prote... 32.0 2.0
mmu:223752 Gramd4, 9930016O13; GRAM domain containing 4 32.0
dre:100332599 mutagen-sensitive 308-like 31.6 3.0
dre:321061 pdcd4b, cb45, pdcd4, sb:cb45, wu:fc84b08; programme... 31.2 3.3
ath:AT3G27040 meprin and TRAF homology domain-containing prote... 30.8 4.9
ath:AT2G01790 meprin and TRAF homology domain-containing prote... 30.4 5.2
hsa:10208 USPL1, C13orf22, D13S106E, DKFZp781K2286, FLJ32952, ... 30.4 6.1
dre:326055 fd60a08; wu:fd60a08 30.4 6.3
dre:100149489 GRAM domain-containing protein 4-like 30.0 6.7
ath:AT5G13210 hypothetical protein 30.0 7.0
xla:100037148 gramd4, dip; GRAM domain containing 4 30.0
hsa:788 SLC25A20, CAC, CACT; solute carrier family 25 (carniti... 29.6 9.2
> tgo:TGME49_060630 DnaJ domain-containing protein ; K09540 translocation
protein SEC63
Length=675
Score = 330 bits (846), Expect = 3e-90, Method: Compositional matrix adjust.
Identities = 161/237 (67%), Positives = 194/237 (81%), Gaps = 0/237 (0%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
K NYEKYGNPDGPG MK+G+GLPRFLVEE Q+ VLS FFLFLLV+LPM+F+CYYQRQKK
Sbjct 188 KANYEKYGNPDGPGNMKVGMGLPRFLVEEKYQLLVLSCFFLFLLVLLPMVFICYYQRQKK 247
Query 63 YAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVVEA 122
YAPNGV+VET+QFLTH M +G+RLKN PE ++ASGESRAM++E++DD +M++I D VE
Sbjct 248 YAPNGVLVETLQFLTHYMAEGSRLKNFPEYLSASGESRAMQVEKEDDVEMREIIDQAVEP 307
Query 123 KKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWY 182
KKR+ P+IVRN YLI HMQRLH LMS RL+ L++LLK SLPITQCM EI VLSDW
Sbjct 308 KKRVLNAPIIVRNYYLILGHMQRLHHLMSDRLRDALDELLKASLPITQCMAEICVLSDWL 367
Query 183 LTADSVLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHCHRGKHVIRELSEFLRQEGE 239
A SVLEFRRCL+Q ++GR+ +L Q+PHF E VRHC RGKH REL +FL+Q+ E
Sbjct 368 HAATSVLEFRRCLVQGLDGRSSTLLQVPHFTLEAVRHCQRGKHAARELGDFLKQDPE 424
> pfa:PF13_0102 DnaJ/SEC63 protein, putative; K09540 translocation
protein SEC63
Length=651
Score = 288 bits (738), Expect = 9e-78, Method: Compositional matrix adjust.
Identities = 126/237 (53%), Positives = 180/237 (75%), Gaps = 0/237 (0%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
K+NYEKYGNPDGPG MK+G+GLP+ L++E Q+ +LS FFL LV +P F+ YYQ+QK+
Sbjct 187 KENYEKYGNPDGPGMMKVGIGLPKLLIDEKYQLLILSIFFLIFLVFIPATFIIYYQKQKQ 246
Query 63 YAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVVEA 122
Y PNGV +ET+Q+LT+ + + +R K+ PE++AA+ ESR +++++DD++ +K + + ++E
Sbjct 247 YGPNGVKIETLQYLTYTINENSRSKSFPEMLAATAESRDIEMKKDDEQYIKTLMEELIEP 306
Query 123 KKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWY 182
KKR F +PVI +N +LI AHMQR H+L+S LKK+L +LK SL IT M+EIS+L DW+
Sbjct 307 KKRTFKIPVITKNYFLILAHMQRRHDLLSEDLKKDLEHILKFSLLITHSMIEISILRDWF 366
Query 183 LTADSVLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHCHRGKHVIRELSEFLRQEGE 239
LTA S L FRRCLIQA + SL QIPHFDE +VRH H+GK ++E+ +F+ Q+ E
Sbjct 367 LTAQSALTFRRCLIQAFEAKNSSLLQIPHFDENIVRHVHKGKFSVKEVLDFVHQDHE 423
> cpv:cgd7_3880 DNAJ domain protein sec63 ortholog, 4 transmembrane
domains ; K09540 translocation protein SEC63
Length=627
Score = 214 bits (545), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 106/239 (44%), Positives = 154/239 (64%), Gaps = 4/239 (1%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
+ NYEKYGNPDGP MK+G+GLP FLV + Q+F+L F L +L V+P+ F+ YY+RQKK
Sbjct 187 RSNYEKYGNPDGPTSMKVGIGLPSFLVSKKYQLFILCFLSLIILFVIPLAFIIYYRRQKK 246
Query 63 YAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIE-RDDDKDMKQICDAVVE 121
YA NGV + T+ F + ++D TR K LPE++A S E R++K +DDK + + + + E
Sbjct 247 YASNGVYLTTLYFYSAAISDSTRFKALPEILALSTEFRSLKKNTSEDDKVISHLANILPE 306
Query 122 AKKRIF--TVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLS 179
KKR F P YLI AH+ R H ++P LKK L +L S+ +T M+EIS+
Sbjct 307 FKKRSFNNNSPSFFTAYYLILAHLYRKHSELTPSLKKVLEDILAKSISLTSSMLEISISR 366
Query 180 DWYLTADSVLEFRRCLIQAINGRTQSLF-QIPHFDEEVVRHCHRGKHVIRELSEFLRQE 237
+++ T+ S+L FRR LI A++G + F QIP+ E V+H +GK +R L EF++Q+
Sbjct 367 NFFHTSTSILAFRRSLIHALDGGPNASFLQIPYITENEVQHIKKGKTAVRNLVEFIKQD 425
> bbo:BBOV_II005390 18.m06448; DnaJ domain containing protein;
K09540 translocation protein SEC63
Length=618
Score = 184 bits (467), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 99/232 (42%), Positives = 153/232 (65%), Gaps = 2/232 (0%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
++NYE+YGNPDGPG MK+G+GLPRFLV+ NQ+ +LS FFL LLVV+P IFL +Y++QK
Sbjct 193 RQNYERYGNPDGPGMMKVGIGLPRFLVDVDNQVLILSLFFLVLLVVIPGIFLYFYRKQKL 252
Query 63 YAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVVEA 122
Y GV +ET+Q + + +++GTR K LPE+ A S E + DD++ +++ +
Sbjct 253 YTSIGVRLETLQLIYYTISEGTRQKALPEIYACSTECASTPATSDDEELLRKFVHDAGDV 312
Query 123 KKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWY 182
K++ T + RN ++ HM R E +SP+L+ ++LK S+ +TQCM+++++ W
Sbjct 313 KRKNVTKEAL-RNFIILLCHMNR-REHLSPKLQAVKAEILKYSVLLTQCMLDVALCRRWL 370
Query 183 LTADSVLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHCHRGKHVIRELSEFL 234
LTA S+++FRRCLIQ I G+ + Q+PH D+E V H R K + E++
Sbjct 371 LTAKSIIDFRRCLIQGITGKRDAFLQVPHIDDECVGHIQRSKAGGKTFGEYV 422
> tpv:TP02_0490 hypothetical protein; K09540 translocation protein
SEC63
Length=659
Score = 182 bits (462), Expect = 8e-46, Method: Compositional matrix adjust.
Identities = 94/233 (40%), Positives = 152/233 (65%), Gaps = 2/233 (0%)
Query 5 NYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKKYA 64
NY KYGNPDGPG MKIG+GLPRFLV+E NQI +LS FFL LL+V P +FL YY+ QK
Sbjct 195 NYAKYGNPDGPGMMKIGIGLPRFLVDENNQIVILSLFFLLLLIVAPSLFLWYYRTQKNLT 254
Query 65 PNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVVEAKK 124
+G+ +ET+Q + + + + TR K+LPE+ + S E + ++ ++++ + + K+
Sbjct 255 ASGIRLETLQLIYYSLNENTRHKSLPEVYSCSSELSNIPYHSSEETELRKYFHILSDYKR 314
Query 125 RIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLT 184
+ + RN L+ H+ R+ +L S +LKK LN++LK S+ IT CM+++++ W LT
Sbjct 315 KNISSETF-RNFLLLLCHLNRIEDL-SSQLKKSLNEVLKYSMLITHCMLDVALSKSWLLT 372
Query 185 ADSVLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHCHRGKHVIRELSEFLRQE 237
SV++FRR ++Q + R++S +QIPHF E + H RGK + + ++++ +
Sbjct 373 FRSVIDFRRGILQGLLTRSESFYQIPHFTEYEINHVGRGKASSKSIEQYVKTD 425
> dre:436861 sec63, zgc:92718; SEC63-like (S. cerevisiae); K09540
translocation protein SEC63
Length=751
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 64/255 (25%), Positives = 124/255 (48%), Gaps = 25/255 (9%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
+KN+E YGNPDGP G+ LP ++V++ N + VL + L +V+LP++ ++ R +
Sbjct 158 RKNWEMYGNPDGPRVTSFGIALPAWIVDQKNSMLVLLVYGLAFMVILPVVVGTWWYRSIR 217
Query 63 YAPNGVMVETVQFLTHVM--TDGTRLKNLPELMAASGE----SRAMKIERDDD-----KD 111
Y+ + +++ T Q H M T +K L ++ A+ E S I R D +
Sbjct 218 YSGDQILINTTQLFMHFMYKTPTMNMKRLVMVLTAAFEFDPRSNKEAIIRPTDNIEVPQL 277
Query 112 MKQICDAVVEAKKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQ- 170
++++ + V+ K+ F P ++ L+ A + R+ +S ++++ ++K + Q
Sbjct 278 IRELGNINVKKKEPPFCYPYSLKARVLLLAQLARMD--VSENIEEDQRFVVKKCPALLQE 335
Query 171 -----CMMEISVLSDWYLTADSVLEFRRCL------IQAINGRTQSLFQIPHFDEEVVRH 219
C + + S L A + C+ +Q + L Q+PHF+EE +R+
Sbjct 336 MINVGCQLTMMATSRGGLRAPRLTSIENCMKLSQMVVQGLQEAKSPLLQLPHFEEEHLRY 395
Query 220 CHRGKHVIRELSEFL 234
C K+ +R L + +
Sbjct 396 CISKKYKVRTLQDLV 410
> dre:100334249 SEC63-like protein-like; K09540 translocation
protein SEC63
Length=751
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 64/255 (25%), Positives = 124/255 (48%), Gaps = 25/255 (9%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
+KN+E YGNPDGP G+ LP ++V++ N + VL + L +V+LP++ ++ R +
Sbjct 158 RKNWEMYGNPDGPRVTSFGIALPAWIVDQKNSMLVLLVYGLAFMVILPVVVGTWWYRSIR 217
Query 63 YAPNGVMVETVQFLTHVM--TDGTRLKNLPELMAASGE----SRAMKIERDDD-----KD 111
Y+ + +++ T Q H M T +K L ++ A+ E S I R D +
Sbjct 218 YSGDQILINTTQLFMHFMYKTPTMNMKRLVMVLTAAFEFDPRSNKEAIIRPTDNIEVPQL 277
Query 112 MKQICDAVVEAKKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQ- 170
++++ + V+ K+ F P ++ L+ A + R+ +S ++++ ++K + Q
Sbjct 278 IRELGNINVKKKEPPFCYPYSLKARVLLLAQLARMD--VSENIEEDQRFVVKKCPALLQE 335
Query 171 -----CMMEISVLSDWYLTADSVLEFRRCL------IQAINGRTQSLFQIPHFDEEVVRH 219
C + + S L A + C+ +Q + L Q+PHF+EE +R+
Sbjct 336 MINVGCQLTMMATSRGGLRAPRLTSIENCMKLSQMVVQGLQEAKSPLLQLPHFEEEHLRY 395
Query 220 CHRGKHVIRELSEFL 234
C K+ +R L + +
Sbjct 396 CISKKYKVRTLQDLV 410
> ath:AT1G79940 ATERDJ2A; heat shock protein binding / unfolded
protein binding; K09540 translocation protein SEC63
Length=687
Score = 91.3 bits (225), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 63/253 (24%), Positives = 125/253 (49%), Gaps = 31/253 (12%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVE-ETNQIFVLSFFFLFLLVVLPMIF-LCYYQRQ 60
++N+EKYG+PDG G ++G+ LP+FL++ + +L + + + ++LP++ + Y R
Sbjct 157 RENFEKYGHPDGRQGFQMGIALPQFLLDIDGASGGILLLWIVGVCILLPLVIAVIYLSRS 216
Query 61 KKYAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAV- 119
KY N VM +T+ ++M + E+ + E + + R DD+ ++++ +V
Sbjct 217 SKYTGNYVMHQTLSAYYYLMKPSLAPSKVMEVFTKAAEYMEIPVRRTDDEPLQKLFMSVR 276
Query 120 -----------VEAKKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPI 168
E K P IV+ LI A + R ++SP L+ + ++L+ + +
Sbjct 277 SELNLDLKNMKQEQAKFWKQHPAIVKTELLIQAQLTRESGVLSPALQGDFRRVLELAPRL 336
Query 169 TQCMMEISVLS------DWYLTADSVLEFRRCLIQAI--NGRTQS---------LFQIPH 211
+ +++++V+ W A V+E +C++QA+ + R S Q+PH
Sbjct 337 LEELLKMAVIPRTAQGHGWLRPAVGVVELSQCIVQAVPLSARKSSGVSSEGISPFLQLPH 396
Query 212 FDEEVVRHCHRGK 224
F + VV+ R K
Sbjct 397 FSDAVVKKIARKK 409
> cel:Y63D3A.6 dnj-29; DNaJ domain (prokaryotic heat shock protein)
family member (dnj-29); K09540 translocation protein SEC63
Length=752
Score = 85.1 bits (209), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 55/242 (22%), Positives = 126/242 (52%), Gaps = 20/242 (8%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
++N+EKYGNPDGP G+ LP++LV + ++VL+F+ L +++LP+ ++ K
Sbjct 161 RENWEKYGNPDGPTATTFGIALPKWLVSKEYGLWVLAFYGLIFMIILPVSVGMWWYSSIK 220
Query 63 YAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGE-------SRAMKIERDDDKD---- 111
Y+ + V+++T + + + R++ + SG ++ ++ DD +
Sbjct 221 YSADKVLLDTTRMYYYFINRTPRMEIGRMIAVLSGSFEFSKQYNKEIQERETDDYEVPRL 280
Query 112 MKQICDAVVEAKKRIFTVPVIVRNCYLINAHMQRLH-ELMSPRLKKELN-------QLLK 163
MKQI + K++ + P +++ L++A++ RL E + L+++ + + ++
Sbjct 281 MKQIAGVNDKGKEQPLSQPYALKSRVLLHAYLSRLPLESQNDALEEDQSYIITRILRFVE 340
Query 164 DSLPITQCMMEISVLSDWYL-TADSVLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHCHR 222
+ + +Q +M S + + T D++L+ + +QA+ + L Q+PH + ++H +
Sbjct 341 EMVNCSQNLMNYSQHTKISIETFDNLLKLQPMFVQALWQKNSPLLQLPHITDYNLQHLRK 400
Query 223 GK 224
+
Sbjct 401 KR 402
> ath:AT4G21180 ATERDJ2B; heat shock protein binding / unfolded
protein binding
Length=661
Score = 79.7 bits (195), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 56/246 (22%), Positives = 118/246 (47%), Gaps = 32/246 (13%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVE---ETNQIFVLSFFFLFLLVVLPMIFLC-YYQ 58
++N+EKYG+PDG G +G+ LP+F++ E+ I +L + L ++LP++ Y
Sbjct 157 RENFEKYGHPDGRQGYTMGIALPQFILNMNGESGGILLLC--TVGLCILLPLVIASIYLW 214
Query 59 RQKKYAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDA 118
R KY N V ++T Q ++ + ++ + E + + + DD+ ++++ +
Sbjct 215 RSSKYTGNHVKLQTRQAYFELLQPSLTPSKVMDIFIRAAEYAEISVRKSDDESLQKLFMS 274
Query 119 VV------------EAKKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSL 166
V E K P ++ LI + R ++SP L+++ +L+ +
Sbjct 275 VKSELNLDPKKLKQEEAKFWKKHPATIKTELLIQKQLTRESSVLSPTLQRDFRHVLEFAP 334
Query 167 PITQCMMEISVLS------DWYLTADSVLEFRRCLIQAI--NGRTQS------LFQIPHF 212
+ + +++++V+ W A V+E +C++QA+ + R S Q+PHF
Sbjct 335 RLLEDLIKMAVIPRNEQGRGWLRPALGVMELSQCIVQAVPLSARKSSSEDIAPFLQLPHF 394
Query 213 DEEVVR 218
+E + +
Sbjct 395 NESIAK 400
> mmu:140740 Sec63, 5730478J10Rik, AI649014, AW319215; SEC63-like
(S. cerevisiae); K09540 translocation protein SEC63
Length=760
Score = 74.3 bits (181), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 55/248 (22%), Positives = 109/248 (43%), Gaps = 29/248 (11%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
+KN+E++GNPDGP G+ LP ++V++ N I VL + L +V+LP++ ++ R +
Sbjct 158 RKNWEEFGNPDGPQATSFGIALPAWIVDQKNSILVLLVYGLAFMVILPVVVGSWWYRSIR 217
Query 63 YAPNGVMVETVQFLTHVM--TDGTRLKNLPELMAASGESRAM----KIERDDDKDM---- 112
Y+ + +++ T Q T+ + T +K L ++A + E R D +
Sbjct 218 YSGDQILIRTTQIYTYFVYKTRNMDMKRLIMVLAGASEFDPQYNKDSTSRPTDNILIPQL 277
Query 113 -KQICDAVVEAKKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLP---- 167
++I ++ + T P ++ L+ +H+ R+ P +E Q + P
Sbjct 278 IREIGSINLKKNEPPLTCPYSLKARVLLLSHLARMK---IPETLEEDQQFMLKKCPALLQ 334
Query 168 -----ITQCMMEISVLSDWYLTADSVLEFRRCL------IQAINGRTQSLFQIPHFDEEV 216
I Q ++ + A ++ C+ +Q + L Q+PH +E+
Sbjct 335 EMVNVICQLIIMARSREEREFRAPTLASLENCMKLSQMAVQGLQQFKSPLLQLPHIEEDN 394
Query 217 VRHCHRGK 224
+R K
Sbjct 395 LRRVSNHK 402
> hsa:11231 SEC63, ERdj2, PRO2507, SEC63L; SEC63 homolog (S. cerevisiae);
K09540 translocation protein SEC63
Length=760
Score = 73.2 bits (178), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 55/248 (22%), Positives = 111/248 (44%), Gaps = 29/248 (11%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
+KN+E++GNPDGP G+ LP ++V++ N I VL + L +V+LP++ ++ R +
Sbjct 158 RKNWEEFGNPDGPQATSFGIALPAWIVDQKNSILVLLVYGLAFMVILPVVVGSWWYRSIR 217
Query 63 YAPNGVMVETVQFLTHVM--TDGTRLKNLPELMAASGE------SRAMKIERDD---DKD 111
Y+ + +++ T Q T+ + T +K L ++A + E A D+ +
Sbjct 218 YSGDQILIRTTQIYTYFVYKTRNMDMKRLIMVLAGASEFDPQYNKDATSRPTDNILIPQL 277
Query 112 MKQICDAVVEAKKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLP---- 167
+++I ++ + T P ++ L+ +H+ R+ P +E Q + P
Sbjct 278 IREIGSINLKKNEPPLTCPYSLKARVLLLSHLARMK---IPETLEEDQQFMLKKCPALLQ 334
Query 168 -----ITQCMMEISVLSDWYLTADSVLEFRRCL------IQAINGRTQSLFQIPHFDEEV 216
I Q ++ + A ++ C+ +Q + L Q+PH +E+
Sbjct 335 EMVNVICQLIVMARNREEREFRAPTLASLENCMKLSQMAVQGLQQFKSPLLQLPHIEEDN 394
Query 217 VRHCHRGK 224
+R K
Sbjct 395 LRRVSNHK 402
> xla:443902 MGC80164 protein; K09540 translocation protein SEC63
Length=755
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 57/247 (23%), Positives = 114/247 (46%), Gaps = 27/247 (10%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
+KN+E++GNPDGP G+ LP ++V++ N + VL + L +V+LP++ ++ + +
Sbjct 158 RKNWEEHGNPDGPQATTFGIALPAWIVDQKNSMLVLLVYGLAFMVILPVVVGSWWYKSIR 217
Query 63 YAPNGVMVETVQFLTHVM--TDGTRLKNLPELMAASGE------SRAMKIERDD---DKD 111
Y+ + +++ T Q T+ + T +K L ++A + E A D+ +
Sbjct 218 YSGDQILIRTTQIYTYFVYKTRNMDMKRLIMVLAGASEFDPQYNKDATSRPADNILIPQL 277
Query 112 MKQICDAVVEAKKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQ- 170
+++I ++ + T P ++ L+ AH+ R+ + LK++ LLK + Q
Sbjct 278 IREIGSINLKKNEPPLTYPYSLKTRVLLLAHLSRMQ--IPETLKEDQQFLLKKCSALLQE 335
Query 171 -----CMMEISVLS--DWYLTADSVLEFRRCL------IQAINGRTQSLFQIPHFDEEVV 217
C + I S + L S+ C+ +Q + L Q+P +E+ +
Sbjct 336 VVNVICQLIIMARSREERELRPPSLSSLENCMKLSQMTVQGLQQFKSPLLQLPFIEEDHL 395
Query 218 RHCHRGK 224
R K
Sbjct 396 RRVSNHK 402
> xla:495416 sec63, MGC132056; SEC63 homolog; K09540 translocation
protein SEC63
Length=754
Score = 70.1 bits (170), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 44/184 (23%), Positives = 94/184 (51%), Gaps = 13/184 (7%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
+KN+E++GNPDGP G+ LP ++V++ N + VL + L +V+LP++ ++ + +
Sbjct 158 RKNWEEHGNPDGPQATTFGIALPAWIVDQKNSMLVLLVYGLAFMVILPVVVGSWWYKSIR 217
Query 63 YAPNGVMVETVQFLTHVM--TDGTRLKNLPELMAASGE------SRAMKIERDD---DKD 111
Y+ + +++ T Q T+ + T +K L ++A + E A D+ +
Sbjct 218 YSGDQILIRTTQIYTYFVYKTRNMDMKRLIMVLAGASEFDPQYNKDATSRPADNILIPQL 277
Query 112 MKQICDAVVEAKKRIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQC 171
+++I ++ + T P ++ L+ AH+ R+ + LK++ +LK + Q
Sbjct 278 IREIGSINLKKNEPPLTCPYSLKTRVLLLAHLSRMQ--IPETLKEDQQFMLKKCSALLQE 335
Query 172 MMEI 175
M+ +
Sbjct 336 MVNV 339
> dre:799690 fb63a09; wu:fb63a09 (EC:3.6.4.13); K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2134
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/150 (21%), Positives = 76/150 (50%), Gaps = 4/150 (2%)
Query 72 TVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVV-EAKKRIFTVP 130
T++ + + T+++ L E+++ + E + + I +D ++Q+ V + F P
Sbjct 1824 TIELFSMSLNAKTKVRGLIEIISNAAEYKNIPIRHHEDTLLRQLAQKVPHKLNNPKFNDP 1883
Query 131 VIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLE 190
+ N L+ AH+ R+ +S L+ + ++L ++ + Q +++ + W A + +E
Sbjct 1884 HVKTNL-LLQAHLSRMQ--LSAELQSDTEEILSKAVRLIQACVDVLSSNGWLSPALAAME 1940
Query 191 FRRCLIQAINGRTQSLFQIPHFDEEVVRHC 220
+ + QA+ + L Q+PHF E+++ C
Sbjct 1941 LAQMVTQAMWSKDSYLRQLPHFTSELIKRC 1970
> sce:YOR254C SEC63, PTL1; Essential subunit of Sec63 complex
(Sec63p, Sec62p, Sec66p and Sec72p); with Sec61 complex, Kar2p/BiP
and Lhs1p forms a channel competent for SRP-dependent
and post-translational SRP-independent protein targeting and
import into the ER; K09540 translocation protein SEC63
Length=663
Score = 53.1 bits (126), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 44/79 (55%), Gaps = 1/79 (1%)
Query 3 KKNYEKYGNPDGPGGMKIGVGLPRFLVEETNQIFVLSFFFLFLLVVLPMIFLCYYQRQKK 62
++NY KYG+PDGP G+ LPRFLV+ + ++ + L ++LP ++ R +
Sbjct 189 RQNYLKYGHPDGPQSTSHGIALPRFLVDGSASPLLVVCYVALLGLILPYFVSRWWARTQS 248
Query 63 YAPNGV-MVETVQFLTHVM 80
Y G+ V F+++++
Sbjct 249 YTKKGIHNVTASNFVSNLV 267
> mmu:320632 Snrnp200, A330064G03Rik, Ascc3l1, BC011390, HELIC2,
KIAA0788, U5-200-KD, U5-200KD; small nuclear ribonucleoprotein
200 (U5) (EC:3.6.4.13); K12854 pre-mRNA-splicing helicase
BRR2 [EC:3.6.4.13]
Length=2136
Score = 53.1 bits (126), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 32/150 (21%), Positives = 74/150 (49%), Gaps = 4/150 (2%)
Query 72 TVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVV-EAKKRIFTVP 130
T++ + + T+++ L E+++ + E + I +D ++Q+ V + F P
Sbjct 1828 TIELFSMSLNAKTKVRGLIEIISNAAEYENIPIRHHEDNLLRQLAQKVPHKLNNPKFNDP 1887
Query 131 VIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLE 190
+ N L+ AH+ R+ +S L+ + ++L ++ + Q +++ + W A + +E
Sbjct 1888 HVKTNL-LLQAHLSRMQ--LSAELQSDTEEILSKAIRLIQACVDVLSSNGWLSPALAAME 1944
Query 191 FRRCLIQAINGRTQSLFQIPHFDEEVVRHC 220
+ + QA+ + L Q+PHF E ++ C
Sbjct 1945 LAQMVTQAMWSKDSYLKQLPHFTSEHIKRC 1974
> hsa:23020 SNRNP200, ASCC3L1, BRR2, FLJ11521, HELIC2, RP33, U5-200KD;
small nuclear ribonucleoprotein 200kDa (U5) (EC:3.6.4.13);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2136
Score = 53.1 bits (126), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 32/150 (21%), Positives = 74/150 (49%), Gaps = 4/150 (2%)
Query 72 TVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVV-EAKKRIFTVP 130
T++ + + T+++ L E+++ + E + I +D ++Q+ V + F P
Sbjct 1828 TIELFSMSLNAKTKVRGLIEIISNAAEYENIPIRHHEDNLLRQLAQKVPHKLNNPKFNDP 1887
Query 131 VIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLE 190
+ N L+ AH+ R+ +S L+ + ++L ++ + Q +++ + W A + +E
Sbjct 1888 HVKTNL-LLQAHLSRMQ--LSAELQSDTEEILSKAIRLIQACVDVLSSNGWLSPALAAME 1944
Query 191 FRRCLIQAINGRTQSLFQIPHFDEEVVRHC 220
+ + QA+ + L Q+PHF E ++ C
Sbjct 1945 LAQMVTQAMWSKDSYLKQLPHFTSEHIKRC 1974
> xla:100158290 snrnp200, ascc3l1; small nuclear ribonucleoprotein
200kDa (U5); K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=457
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 32/150 (21%), Positives = 74/150 (49%), Gaps = 4/150 (2%)
Query 72 TVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVVEA-KKRIFTVP 130
T++ + + T+++ L E+++ + E ++ I +D ++Q+ V F P
Sbjct 149 TIELFSMSLNAKTKVRGLIEIISNAAEYESIPIRHHEDNLLRQLAQKVPHKLTNPKFNDP 208
Query 131 VIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLE 190
+ N L+ AH+ R+ +S L+ + ++L ++ + Q +++ + W A + +E
Sbjct 209 HVKTNL-LLQAHLSRMQ--LSAELQSDTEEILGKAVRLIQACVDVLSSNGWLSPALAAME 265
Query 191 FRRCLIQAINGRTQSLFQIPHFDEEVVRHC 220
+ + QA+ + L Q+PHF E ++ C
Sbjct 266 LAQMVTQAMWSKDSYLRQLPHFSSEHIKRC 295
> tgo:TGME49_023390 sec63 domain-containing DEAD/DEAH box helicase,
putative ; K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2198
Score = 45.8 bits (107), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 34/156 (21%), Positives = 71/156 (45%), Gaps = 6/156 (3%)
Query 68 VMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAV---VEAKK 124
V V T++ +T + + L E++AAS E + + ++ +K + + + A
Sbjct 1887 VKVNTIELFNRSLTPTCKRRALLEILAASSEFSTLPLRPGEEGTLKGLAQRLGVRLPANS 1946
Query 125 RIFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLT 184
P + L+ AH R + L + LL+ S+ + ++++ + W +
Sbjct 1947 EDLNKPS-TKALILLYAHFNRTP--LPSDLIADQKVLLEPSIRLLHALVDVISSNGWLVP 2003
Query 185 ADSVLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHC 220
A S +E + ++QA+ +L Q+PHF +E+V
Sbjct 2004 ALSAMEICQAVVQAMTTACSALKQLPHFTDELVEQA 2039
> hsa:652147 u5 small nuclear ribonucleoprotein 200 kDa helicase-like
Length=1700
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/150 (19%), Positives = 71/150 (47%), Gaps = 4/150 (2%)
Query 72 TVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVV-EAKKRIFTVP 130
T++ + + T+++ L E+++ + E + I +D ++Q+ V + F P
Sbjct 1392 TIELFSMSLNAKTKVRGLIEIISNAAEYENIPIRHHEDNLLRQLAQKVPHKLNNPKFNDP 1451
Query 131 VIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLE 190
+ N L+ AH+ R+ +S L+ + ++L ++ + Q +++ + W A + +E
Sbjct 1452 HVKTNL-LLQAHLSRMQ--LSAELQSDTEEILSKAIRLIQACVDVLSSNGWLSPALAAME 1508
Query 191 FRRCLIQAINGRTQSLFQIPHFDEEVVRHC 220
+ + QA+ L ++P F + + C
Sbjct 1509 LAQMVTQAMWSEDSYLRRLPPFPSGLFKRC 1538
> sce:YER172C BRR2, PRP44, RSS1, SLT22, SNU246; Brr2p (EC:3.6.1.-);
K12854 pre-mRNA-splicing helicase BRR2 [EC:3.6.4.13]
Length=2163
Score = 43.9 bits (102), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 31/155 (20%), Positives = 77/155 (49%), Gaps = 4/155 (2%)
Query 67 GVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAV-VEAKKR 125
GV T+Q +++ + LKN+ +++ + E ++ + + D + ++ + + +
Sbjct 1857 GVSFFTIQSFVSSLSNTSTLKNMLYVLSTAVEFESVPLRKGDRALLVKLSKRLPLRFPEH 1916
Query 126 IFTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTA 185
+ V + L+ A+ RL + + +L +L+ +P+ +++I + ++ YL A
Sbjct 1917 TSSGSVSFKVFLLLQAYFSRLE--LPVDFQNDLKDILEKVVPLINVVVDI-LSANGYLNA 1973
Query 186 DSVLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHC 220
+ ++ + LIQ + L QIPHF+ +++ C
Sbjct 1974 TTAMDLAQMLIQGVWDVDNPLRQIPHFNNKILEKC 2008
> cel:Y46G5A.4 hypothetical protein; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2145
Score = 43.5 bits (101), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 32/145 (22%), Positives = 73/145 (50%), Gaps = 4/145 (2%)
Query 68 VMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVV-EAKKRI 126
+ +T++ + + + T+ + L E+++AS E + + +D ++Q+ + + + K +
Sbjct 1823 ISYQTIELFSMSLKEKTKTRALIEIISASSEFGNVPMRHKEDVILRQLAERLPGQLKNQK 1882
Query 127 FTVPVIVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTAD 186
FT P + N LI+AH+ R+ ++ L K+ ++ + + Q +++ + W A
Sbjct 1883 FTDPHVKVNL-LIHAHLSRVK--LTAELNKDTELIVLRACRLVQACVDVLSSNGWLSPAI 1939
Query 187 SVLEFRRCLIQAINGRTQSLFQIPH 211
+E + L QA+ L Q+PH
Sbjct 1940 HAMELSQMLTQAMYSNEPYLKQLPH 1964
> ath:AT1G20960 emb1507 (embryo defective 1507); ATP binding /
ATP-dependent helicase/ helicase/ nucleic acid binding / nucleoside-triphosphatase/
nucleotide binding; K12854 pre-mRNA-splicing
helicase BRR2 [EC:3.6.4.13]
Length=2171
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 31/161 (19%), Positives = 76/161 (47%), Gaps = 12/161 (7%)
Query 72 TVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVVEAKKRIFTVPV 131
T++ + +++ T++K L E++ ++ E + I ++ ++++ + ++ F P
Sbjct 1855 TIERFSSLLSSKTKMKGLLEILTSASEYDMIPIRPGEEDTVRRL----INHQRFSFENPK 1910
Query 132 I----VRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADS 187
V+ L+ AH R + + L + +L + + Q M+++ + W A
Sbjct 1911 CTDPHVKANALLQAHFSRQN--IGGNLAMDQRDVLLSATRLLQAMVDVISSNGWLNLALL 1968
Query 188 VLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHCHR--GKHV 226
+E + + Q + R L Q+PHF +++ + C GK++
Sbjct 1969 AMEVSQMVTQGMWERDSMLLQLPHFTKDLAKRCQENPGKNI 2009
> ath:AT2G42270 U5 small nuclear ribonucleoprotein helicase, putative
Length=2172
Score = 39.3 bits (90), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 28/150 (18%), Positives = 71/150 (47%), Gaps = 2/150 (1%)
Query 72 TVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVVEAKKRIFTVPV 131
T++ + ++ T++K L E++ ++ E + I ++ ++++ + + +
Sbjct 1856 TIERFSSLLASKTKMKGLLEILTSASEYDLIPIRPGEEDAVRRLINHQRFSFQNPRCTDP 1915
Query 132 IVRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEF 191
V+ L+ AH R + +S L + ++L + + Q M+++ + A +E
Sbjct 1916 RVKTSALLQAHFSR--QKISGNLVMDQCEVLLSATRLLQAMVDVISSNGCLNLALLAMEV 1973
Query 192 RRCLIQAINGRTQSLFQIPHFDEEVVRHCH 221
+ + Q + R L Q+PHF +++ + CH
Sbjct 1974 SQMVTQGMWDRDSMLLQLPHFTKDLAKRCH 2003
> cel:Y54E2A.6 hypothetical protein; K01529 [EC:3.6.1.-]
Length=1798
Score = 34.7 bits (78), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 33/158 (20%), Positives = 67/158 (42%), Gaps = 12/158 (7%)
Query 71 ETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQICDAVVEAKKRI-FTV 129
ETV+FL + G ++N+ +++ E + + ++D + + ++ K RI F+
Sbjct 1464 ETVRFLVKSLHSGCSVENMLKILTDVPEYAEIPVRHNED-----LINTELQKKLRIRFST 1518
Query 130 PVIVRNC----YLINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTA 185
V+ + L AH R ++ + +L +L + I Q M E++ L +W
Sbjct 1519 SVMGTSACKAHLLFQAHFMR--TVLPTDYRTDLKSVLDQCIRILQAMREMARLKNWLSAT 1576
Query 186 DSVLEFRRCLIQAINGRTQSLFQIPHFDEEVVRHCHRG 223
+++ ++ A L +PH E R G
Sbjct 1577 MNIILLQQMCHSARWHDDHPLLCLPHLSHEDARSIGDG 1614
> hsa:23151 GRAMD4, DIP, KIAA0767, MGC149847, MGC90497, dA59H18.1,
dJ439F8.1; GRAM domain containing 4
Length=578
Score = 33.1 bits (74), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 17/59 (28%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 149 LMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEFRRCLIQAINGRTQSLF 207
L + RL + + +L + + P+T + +S LSDWY S + F + +G LF
Sbjct 196 LSARRLTENMRRLKRGAKPVTNFVKNLSALSDWYSVYTSAIAFTVYMNAVWHGWAIPLF 254
> mmu:77987 Ascc3, ASC1p200, B630009I04Rik, BC023451, D430001L07Rik,
D630041L21, Helic1, RNAH; activating signal cointegrator
1 complex subunit 3; K01529 [EC:3.6.1.-]
Length=2198
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 39/84 (46%), Gaps = 4/84 (4%)
Query 138 LINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEFRRCLIQ 197
L+ AH+ R L P + +L +L + Q M++++ W +T ++ + +IQ
Sbjct 1895 LLQAHLSRAM-LPCPDYDTDTKTVLDQALRVCQAMLDVAASQGWLVTVLNITHLIQMVIQ 1953
Query 198 AINGRTQSLFQIPHFDEEVVRHCH 221
+ SL IP+ ++ H H
Sbjct 1954 GRWLKDSSLLTIPNIEQ---HHLH 1974
> pfa:PFD1060w U5 small nuclear ribonucleoprotein-specific protein,
putative; K01509 adenosinetriphosphatase [EC:3.6.1.3]
Length=2874
Score = 32.0 bits (71), Expect = 2.0, Method: Composition-based stats.
Identities = 41/172 (23%), Positives = 76/172 (44%), Gaps = 22/172 (12%)
Query 52 IFLCYYQRQKKYAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKD 111
IFLC Q A N V V T +FL + + LKN A+ E K + +D
Sbjct 2559 IFLCLKIAQ---ACNNVQV-TYEFLKLSINNENTLKN------ANIEDNLNKDTKSEDYK 2608
Query 112 MKQICDAVVEAKKRIFTVPVI----VRNCYLINAHMQRLHELMSPRLKKELNQLLKDSLP 167
Q + + + +VP+ ++ L+ AH+ R + +E +L +
Sbjct 2609 KDQYINLL-----QFMSVPMYFTSHLKAFILLQAHIHRYS--LPLNYIQETKTVLLKAYK 2661
Query 168 ITQCMMEISVLSDWYLTADSVLEFRRCLIQAINGRTQS-LFQIPHFDEEVVR 218
+ ++++ ++ V+E + L Q++ QS L+Q+PHFDE +++
Sbjct 2662 LINSLIDVISSNNILNFCLFVMEVSQMLTQSMKSTDQSNLYQLPHFDEHLIK 2713
> mmu:223752 Gramd4, 9930016O13; GRAM domain containing 4
Length=627
Score = 32.0 bits (71), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 149 LMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEF 191
L + RL + + +L + + P+T + +S LSDWY S + F
Sbjct 226 LSARRLTENMRRLKRGAKPVTNFVKNLSALSDWYSIYTSAIAF 268
> dre:100332599 mutagen-sensitive 308-like
Length=864
Score = 31.6 bits (70), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 19/78 (24%), Positives = 37/78 (47%), Gaps = 1/78 (1%)
Query 138 LINAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEFRRCLIQ 197
L+ AH R +L + +L +++ I Q M+++ W ++A S+ + +IQ
Sbjct 728 LLQAHFSRA-QLPCSDYGTDTKTVLDNAIRICQAMLDVVANEGWLVSALSLCNLVQMIIQ 786
Query 198 AINGRTQSLFQIPHFDEE 215
A SL +PH ++
Sbjct 787 ARWLHDSSLLTLPHIQKQ 804
> dre:321061 pdcd4b, cb45, pdcd4, sb:cb45, wu:fc84b08; programmed
cell death 4b
Length=470
Score = 31.2 bits (69), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 26/51 (50%), Gaps = 9/51 (17%)
Query 167 PITQCMMEISVLSDWYLTADSVLEFRRCLIQAINGRTQSLFQIPHFDEEVV 217
P+TQ + E+++L YL + +E RCL + ++PHF E V
Sbjct 324 PVTQLIKEVNLLLKEYLLSGDTVEAERCLRE---------LEVPHFHHEFV 365
> ath:AT3G27040 meprin and TRAF homology domain-containing protein
/ MATH domain-containing protein
Length=358
Score = 30.8 bits (68), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 42/82 (51%), Gaps = 8/82 (9%)
Query 44 FLLVVLPMI-FLCYYQRQKKYAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAM 102
FL VVL M LC + + +G + E L V G +L L + + +G+SR
Sbjct 278 FLDVVLSMTEILCKFPEE---LSSGDLAEAYSALRFVTKAGFKLDWLEKKLKETGKSRLQ 334
Query 103 KIERDDDKDMKQIC---DAVVE 121
+IE +D KD+K C DA+++
Sbjct 335 EIE-EDLKDLKVKCADMDALLD 355
> ath:AT2G01790 meprin and TRAF homology domain-containing protein
/ MATH domain-containing protein
Length=269
Score = 30.4 bits (67), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 34/66 (51%), Gaps = 4/66 (6%)
Query 59 RQKKYAPNGVMVETVQFLTHVMTDGTRLKNLPELMAASGESRAMKIERDDDKDMKQIC-- 116
+ K NG + E L V G +L L + + +G+SR +IE +D KD+K C
Sbjct 202 KSTKELSNGDLAEAYSALRFVTKAGFKLDWLEKKLKETGKSRLQEIE-EDLKDLKVKCAD 260
Query 117 -DAVVE 121
DA++E
Sbjct 261 MDALLE 266
> hsa:10208 USPL1, C13orf22, D13S106E, DKFZp781K2286, FLJ32952,
RP11-121O19.1, bA121O19.1; ubiquitin specific peptidase like
1
Length=1092
Score = 30.4 bits (67), Expect = 6.1, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 5/75 (6%)
Query 140 NAHMQRLHELMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEFRRCLIQAI 199
N H+++ H SP KK ++ DSL C+ +++ S + D EF A+
Sbjct 1017 NTHLRQDHNYCSPT-KKNPCEVQPDSLTNNACVRTLNLESP--MKTDIFDEFFSS--SAL 1071
Query 200 NGRTQSLFQIPHFDE 214
N +PHFDE
Sbjct 1072 NALANDTLDLPHFDE 1086
> dre:326055 fd60a08; wu:fd60a08
Length=585
Score = 30.4 bits (67), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 149 LMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEFRRCLIQAINGRTQSLF 207
L + RL + + +L + + P+T M +S LS W+ S + F + A +G +F
Sbjct 196 LSAKRLTENMRRLKRGAKPVTNFMRNLSALSSWHSVYTSAIAFIIYMNAAWHGWVLPMF 254
> dre:100149489 GRAM domain-containing protein 4-like
Length=574
Score = 30.0 bits (66), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 30/59 (50%), Gaps = 0/59 (0%)
Query 149 LMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEFRRCLIQAINGRTQSLF 207
L + RL + + +L + + P+T + +S LS+W+ S + F + A +G LF
Sbjct 192 LSAKRLTENMRRLKRGARPVTNFLRNLSALSNWHSVYTSAIAFIIYMNAAWHGWAIPLF 250
> ath:AT5G13210 hypothetical protein
Length=673
Score = 30.0 bits (66), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 28/125 (22%), Positives = 55/125 (44%), Gaps = 10/125 (8%)
Query 99 SRAMKIERDDDKDMKQICDAVVEAKKRIFTVPVIVRNCYLINAHMQRLHE----LMSPRL 154
+RA+++ + K+ + A ++ KK+ ++ Y + + LHE L + +L
Sbjct 218 TRALRVANAERKNQAEKARASLDRKKKKVSMGKDAFTRYSCDPDYRYLHERVSDLFANQL 277
Query 155 KKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEFRRCLIQAINGRTQSLFQIPHFDE 214
KK+L L D EIS+ + W + DS + L ++I + + P ++
Sbjct 278 KKDLEFLTSDK------PNEISLAAKWCPSLDSSFDKATLLCESIARKIFTRESFPEYEG 331
Query 215 EVVRH 219
V H
Sbjct 332 VVEAH 336
> xla:100037148 gramd4, dip; GRAM domain containing 4
Length=578
Score = 30.0 bits (66), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 149 LMSPRLKKELNQLLKDSLPITQCMMEISVLSDWYLTADSVLEF 191
L + RL + + +L + + P+T + +S LSDW+ S + F
Sbjct 196 LSARRLTENMRRLKRGAKPVTNFVKNLSALSDWHSVYTSAIAF 238
> hsa:788 SLC25A20, CAC, CACT; solute carrier family 25 (carnitine/acylcarnitine
translocase), member 20; K15109 solute carrier
family 25 (mitochondrial carnitine/acylcarnitine transporter),
member 20/29
Length=301
Score = 29.6 bits (65), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 75 FLTHVMTDGTRLKNLPELMAASGESR 100
F T +MT G R+K L ++ A+SGES+
Sbjct 123 FTTGIMTPGERIKCLLQIQASSGESK 148
Lambda K H
0.325 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8338372120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40