bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4511_orf2
Length=100
Score E
Sequences producing significant alignments: (Bits) Value
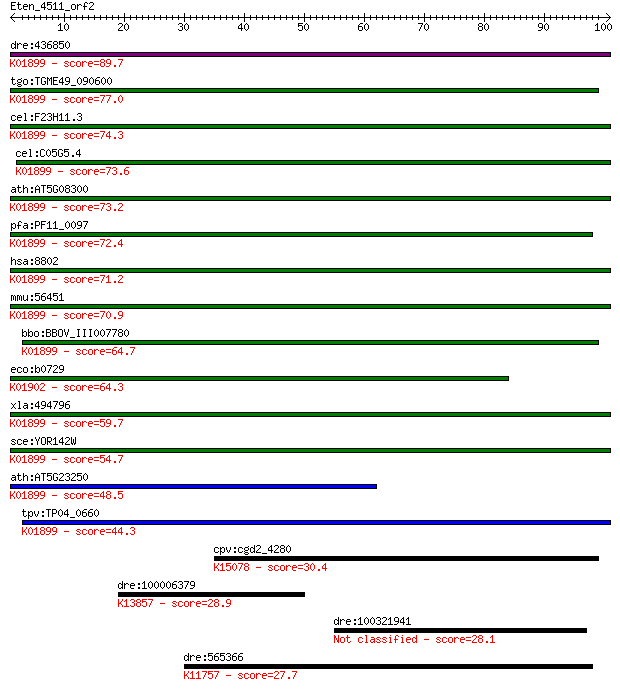
dre:436850 suclg1, MGC103729, zgc:103729, zgc:92738; succinate... 89.7 2e-18
tgo:TGME49_090600 succinyl-CoA ligase alpha subunit, putative ... 77.0 1e-14
cel:F23H11.3 hypothetical protein; K01899 succinyl-CoA synthet... 74.3 9e-14
cel:C05G5.4 hypothetical protein; K01899 succinyl-CoA syntheta... 73.6 2e-13
ath:AT5G08300 succinyl-CoA ligase (GDP-forming) alpha-chain, m... 73.2 2e-13
pfa:PF11_0097 succinyl-CoA synthetase alpha subunit, putative ... 72.4 4e-13
hsa:8802 SUCLG1, FLJ21114, FLJ43513, GALPHA, MTDPS9, SUCLA1; s... 71.2 8e-13
mmu:56451 Suclg1, 1500000I01Rik, Sucla1; succinate-CoA ligase,... 70.9 9e-13
bbo:BBOV_III007780 17.m07682; succinyl-CoA synthetase, alpha s... 64.7 7e-11
eco:b0729 sucD, ECK0717, JW0718; succinyl-CoA synthetase, NAD(... 64.3 1e-10
xla:494796 suclg1, galpha, mtdps9, sucla1; succinate-CoA ligas... 59.7 2e-09
sce:YOR142W LSC1; Alpha subunit of succinyl-CoA ligase, which ... 54.7 7e-08
ath:AT5G23250 succinyl-CoA ligase (GDP-forming) alpha-chain, m... 48.5 5e-06
tpv:TP04_0660 succinyl-CoA synthetase subunit alpha (EC:6.2.1.... 44.3 1e-04
cpv:cgd2_4280 holiday junction resolvase, S1x1p, URI domain nu... 30.4 1.4
dre:100006379 slc4a5; solute carrier family 4, sodium bicarbon... 28.9 4.3
dre:100321941 bag6l, bat3l; BCL2-associated athanogene 6, like 28.1 7.2
dre:565366 pbrm1; polybromo 1; K11757 protein polybromo-1 27.7 9.4
> dre:436850 suclg1, MGC103729, zgc:103729, zgc:92738; succinate-CoA
ligase, GDP-forming, alpha subunit (EC:6.2.1.4); K01899
succinyl-CoA synthetase alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=324
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 56/100 (56%), Positives = 72/100 (72%), Gaps = 0/100 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E+FL+DP EGI LIGE GG++E+ AAE+LK HN G+ AKP SFI GL APP ++ G
Sbjct 219 EVFLQDPKTEGIILIGEIGGDAEENAAEYLKQHNSGANAKPVVSFIAGLTAPPGRRMGHA 278
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GA GGKGG KEKI +AGV+V++ P Q+G+ +FK F
Sbjct 279 GAFIAGGKGGAKEKIAALQSAGVVVSMSPAQLGSTIFKEF 318
> tgo:TGME49_090600 succinyl-CoA ligase alpha subunit, putative
(EC:6.2.1.4); K01899 succinyl-CoA synthetase alpha subunit
[EC:6.2.1.4 6.2.1.5]
Length=333
Score = 77.0 bits (188), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 45/98 (45%), Positives = 60/98 (61%), Gaps = 2/98 (2%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E F+ DP +GI LIGE GG++E++AA +LK HN + KP SFI GL APP ++ G
Sbjct 230 ERFVNDPETKGIVLIGEIGGSAEEEAAAWLKEHN--KEQKPVVSFIAGLTAPPGRRMGHA 287
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFK 98
GAI GGKG + KI +AGV V P +G + +
Sbjct 288 GAIVSGGKGTAEGKIAALEDAGVHVVRNPAAMGQKMLE 325
> cel:F23H11.3 hypothetical protein; K01899 succinyl-CoA synthetase
alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=321
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 49/100 (49%), Positives = 63/100 (63%), Gaps = 0/100 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E+FL+D +GI LIGE GG +E++AAEFLK N GS AKP SFI G+ APP ++ G
Sbjct 220 EVFLEDEQTKGIILIGEIGGQAEEQAAEFLKSRNSGSNAKPVVSFIAGVTAPPGRRMGHA 279
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKG +KI NA V+V P ++G + K
Sbjct 280 GAIIAGGKGTAGDKIEALRNANVVVTDSPAKLGVAMQKAL 319
> cel:C05G5.4 hypothetical protein; K01899 succinyl-CoA synthetase
alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=322
Score = 73.6 bits (179), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 47/99 (47%), Positives = 64/99 (64%), Gaps = 0/99 (0%)
Query 2 IFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSPG 61
+FL+DP +GI LIGE GG++E++AA +LK HN G+ KP SFI G+ APP ++ G G
Sbjct 220 VFLEDPETKGIILIGEIGGSAEEEAAAYLKEHNSGANRKPVVSFIAGVTAPPGRRMGHAG 279
Query 62 AIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
AI GGKG +KI AGV+V P ++GT + F
Sbjct 280 AIISGGKGTAADKINALREAGVVVTDSPAKLGTSMATAF 318
> ath:AT5G08300 succinyl-CoA ligase (GDP-forming) alpha-chain,
mitochondrial, putative / succinyl-CoA synthetase, alpha chain,
putative / SCS-alpha, putative; K01899 succinyl-CoA synthetase
alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=347
Score = 73.2 bits (178), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 43/100 (43%), Positives = 58/100 (58%), Gaps = 3/100 (3%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E F DP EGI LIGE GG +E+ AA +K KP +FI GL APP ++ G
Sbjct 243 EKFFVDPQTEGIVLIGEIGGTAEEDAAALIKASG---TEKPVVAFIAGLTAPPGRRMGHA 299
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKG ++KI +AGV V P ++G+ +++ F
Sbjct 300 GAIVSGGKGTAQDKIKSLNDAGVKVVESPAKIGSAMYELF 339
> pfa:PF11_0097 succinyl-CoA synthetase alpha subunit, putative
(EC:6.2.1.4); K01899 succinyl-CoA synthetase alpha subunit
[EC:6.2.1.4 6.2.1.5]
Length=327
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 41/100 (41%), Positives = 61/100 (61%), Gaps = 3/100 (3%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGS---KAKPGGSFIGGLKAPPRQKK 57
++FL+D E I LIGE GG+SE++ A++L +N+ + K KP +FI G APP ++
Sbjct 221 KLFLEDDQTECILLIGEIGGDSEEQVAQWLIQNNVDNGKRKKKPVFAFIAGKCAPPGKRM 280
Query 58 GSPGAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLF 97
G GA+ GGKG KI NAGV V + P ++G ++
Sbjct 281 GHAGALISGGKGTADGKIEALQNAGVHVILNPTKMGEDIY 320
> hsa:8802 SUCLG1, FLJ21114, FLJ43513, GALPHA, MTDPS9, SUCLA1;
succinate-CoA ligase, alpha subunit (EC:6.2.1.4); K01899 succinyl-CoA
synthetase alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=346
Score = 71.2 bits (173), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 59/100 (59%), Positives = 71/100 (71%), Gaps = 0/100 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
EIFL D A EGI LIGE GGN+E+ AAEFLK HN G +KP SFI GL APP ++ G
Sbjct 241 EIFLNDSATEGIILIGEIGGNAEENAAEFLKQHNSGPNSKPVVSFIAGLTAPPGRRMGHA 300
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKGG KEKI +AGV+V++ P Q+GT ++K F
Sbjct 301 GAIIAGGKGGAKEKISALQSAGVVVSMSPAQLGTTIYKEF 340
> mmu:56451 Suclg1, 1500000I01Rik, Sucla1; succinate-CoA ligase,
GDP-forming, alpha subunit (EC:6.2.1.4); K01899 succinyl-CoA
synthetase alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=346
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 58/100 (58%), Positives = 72/100 (72%), Gaps = 0/100 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E+FL DPA EGI LIGE GG++E+ AA FLK HN G KAKP SFI G+ APP ++ G
Sbjct 241 EVFLNDPATEGIILIGEIGGHAEENAAAFLKEHNSGPKAKPVVSFIAGITAPPGRRMGHA 300
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKGG KEKI +AGV+V++ P Q+GT ++K F
Sbjct 301 GAIIAGGKGGAKEKISALQSAGVVVSMSPAQLGTTIYKEF 340
> bbo:BBOV_III007780 17.m07682; succinyl-CoA synthetase, alpha
subunit family protein (EC:6.2.1.4); K01899 succinyl-CoA synthetase
alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=328
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 39/96 (40%), Positives = 55/96 (57%), Gaps = 3/96 (3%)
Query 3 FLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSPGA 62
F+ DP +GI LIGE GGN+E+ AAE++K + KP + I G APP ++ G GA
Sbjct 232 FVDDPQTKGIMLIGEIGGNAEEDAAEWIKAN---PTDKPIVALIAGTCAPPGRRMGHAGA 288
Query 63 IFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFK 98
I G G + KI +AGV+V P +G+ + K
Sbjct 289 IISGNTGTAQGKIKALRDAGVVVAETPALLGSTMKK 324
> eco:b0729 sucD, ECK0717, JW0718; succinyl-CoA synthetase, NAD(P)-binding,
alpha subunit (EC:6.2.1.5); K01902 succinyl-CoA
synthetase alpha subunit [EC:6.2.1.5]
Length=289
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/83 (43%), Positives = 48/83 (57%), Gaps = 4/83 (4%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E+F KDP E I +IGE GG++E++AA ++K H KP +I G+ AP ++ G
Sbjct 193 EMFEKDPQTEAIVMIGEIGGSAEEEAAAYIKEH----VTKPVVGYIAGVTAPKGKRMGHA 248
Query 61 GAIFFGGKGGGKEKIPPPPNAGV 83
GAI GGKG EK AGV
Sbjct 249 GAIIAGGKGTADEKFAALEAAGV 271
> xla:494796 suclg1, galpha, mtdps9, sucla1; succinate-CoA ligase,
alpha subunit (EC:6.2.1.4); K01899 succinyl-CoA synthetase
alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=195
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 54/100 (54%), Positives = 68/100 (68%), Gaps = 0/100 (0%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
++FL DP EGI LIGE GG+ E+ AAEFL+ +N G AKP SFI GL APP ++ G
Sbjct 90 DVFLSDPKTEGIILIGEIGGSGEENAAEFLRVNNSGPNAKPVVSFIAGLTAPPGRRMGHA 149
Query 61 GAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
GAI GGKGG KEKI AGV+V++ P Q+G+ + K F
Sbjct 150 GAIIAGGKGGAKEKIAALQKAGVVVSMSPAQLGSTMHKEF 189
> sce:YOR142W LSC1; Alpha subunit of succinyl-CoA ligase, which
is a mitochondrial enzyme of the TCA cycle that catalyzes
the nucleotide-dependent conversion of succinyl-CoA to succinate;
phosphorylated (EC:6.2.1.4); K01899 succinyl-CoA synthetase
alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=329
Score = 54.7 bits (130), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 52/103 (50%), Gaps = 3/103 (2%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLG-SKAKPGGSFIGGLKAPPRQ--KK 57
++FL+D EGI ++GE GG +E +AA+FLK +N SK P SFI G A + +
Sbjct 223 KLFLEDETTEGIIMLGEIGGKAEIEAAQFLKEYNFSRSKPMPVASFIAGTVAGQMKGVRM 282
Query 58 GSPGAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
G GAI G + K + GV V P +G L F
Sbjct 283 GHSGAIVEGSGTDAESKKQALRDVGVAVVESPGYLGQALLDQF 325
> ath:AT5G23250 succinyl-CoA ligase (GDP-forming) alpha-chain,
mitochondrial, putative / succinyl-CoA synthetase, alpha chain,
putative / SCS-alpha, putative; K01899 succinyl-CoA synthetase
alpha subunit [EC:6.2.1.4 6.2.1.5]
Length=297
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 28/61 (45%), Positives = 35/61 (57%), Gaps = 3/61 (4%)
Query 1 EIFLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSP 60
E F DP EGI LIGE GG +E+ AA +K + KP +FI GL APP ++ G
Sbjct 238 EKFFVDPQTEGIVLIGEIGGTAEEDAAALIKENGTD---KPVVAFIAGLTAPPGRRMGHA 294
Query 61 G 61
G
Sbjct 295 G 295
> tpv:TP04_0660 succinyl-CoA synthetase subunit alpha (EC:6.2.1.4);
K01899 succinyl-CoA synthetase alpha subunit [EC:6.2.1.4
6.2.1.5]
Length=329
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 55/98 (56%), Gaps = 3/98 (3%)
Query 3 FLKDPAPEGIFLIGENGGNSEKKAAEFLKPHNLGSKAKPGGSFIGGLKAPPRQKKGSPGA 62
F+K+P +GI +IGE GG +E+ A+++K + KP + I G+ APP ++ G GA
Sbjct 232 FVKNPETKGILIIGEIGGTAEEDVADWIKAN---PTDKPVVAIIAGISAPPGKRMGHAGA 288
Query 63 IFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGTPLFKGF 100
I G KG KI +AGV+V P ++G +F
Sbjct 289 IISGNKGTAHGKIEALRDAGVVVPNSPAEMGLAMFNAL 326
> cpv:cgd2_4280 holiday junction resolvase, S1x1p, URI domain
nuclease ; K15078 structure-specific endonuclease subunit SLX1
[EC:3.6.1.-]
Length=410
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 23/64 (35%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Query 35 LGSKAKPGGSFIGGLKAPPRQKKGSPGAIFFGGKGGGKEKIPPPPNAGVLVNIFPPQVGT 94
L S+AK S+IG P R+ + G I KG K K P N G+ V FP +V
Sbjct 13 LLSEAKKKASYIGYSVNPCRRLRQHNGEI---KKGAKKTKSGVPWNLGICVGGFPDRVAA 69
Query 95 PLFK 98
F+
Sbjct 70 LRFE 73
> dre:100006379 slc4a5; solute carrier family 4, sodium bicarbonate
cotransporter, member 5; K13857 solute carrier family
4 (sodium bicarbonate cotransporter), member 5
Length=1067
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 19 GGNSEKKAAEFLKPHNLGSKAKPGGSFIGGL 49
GG++ + E PH LG + K G F GGL
Sbjct 445 GGSASAEDEEMPAPHELGEELKFTGRFCGGL 475
> dre:100321941 bag6l, bat3l; BCL2-associated athanogene 6, like
Length=1027
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 6/44 (13%)
Query 55 QKKGSPGAIFFGGKGGGKEKIPPPPNAGVLVN--IFPPQVGTPL 96
Q++G+ GA G G+ +PPPP A V++ P+V P+
Sbjct 352 QQQGTAGA----GAQNGESPVPPPPQARVVITRPTLSPRVPQPM 391
> dre:565366 pbrm1; polybromo 1; K11757 protein polybromo-1
Length=1581
Score = 27.7 bits (60), Expect = 9.4, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 32/76 (42%), Gaps = 9/76 (11%)
Query 30 LKPHNLGSKAKPGGSFIGGLKAPPRQ-----KKGSPGAIF---FGGKGGGKEKIPPPPNA 81
+ PH +G A P +F GL P GS G+ + GG G++ PP P
Sbjct 1412 MPPHPMGLPALPL-NFPWGLPGMPFHGMNGMAPGSTGSAYGTQMGGMNSGQQVPPPYPGQ 1470
Query 82 GVLVNIFPPQVGTPLF 97
G + Q TP+F
Sbjct 1471 GQVGQPAHQQPSTPMF 1486
Lambda K H
0.315 0.145 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2041372988
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40