bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4493_orf2
Length=62
Score E
Sequences producing significant alignments: (Bits) Value
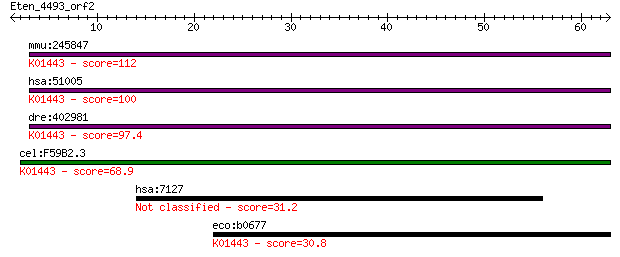
mmu:245847 Amdhd2, 5730457F11Rik; amidohydrolase domain contai... 112 2e-25
hsa:51005 AMDHD2; amidohydrolase domain containing 2 (EC:3.5.1... 100 1e-21
dre:402981 MGC77775, amdhd2; zgc:77775 (EC:3.5.1.25); K01443 N... 97.4 1e-20
cel:F59B2.3 hypothetical protein; K01443 N-acetylglucosamine-6... 68.9 3e-12
hsa:7127 TNFAIP2, B94, EXOC3L3; tumor necrosis factor, alpha-i... 31.2 0.84
eco:b0677 nagA, ECK0665, JW0663; N-acetylglucosamine-6-phospha... 30.8 1.0
> mmu:245847 Amdhd2, 5730457F11Rik; amidohydrolase domain containing
2 (EC:3.5.1.25); K01443 N-acetylglucosamine-6-phosphate
deacetylase [EC:3.5.1.25]
Length=409
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/60 (90%), Positives = 56/60 (93%), Gaps = 0/60 (0%)
Query 3 LGQQEVEVDGLTAYVAGTNTLSGSIAPMDTCVRHFLQATGCSVESALEAASLHPAQLLGL 62
LGQQEVEVDGL AY+AGT TL GSIAPMD CVRHFLQATGCSVESALEAASLHPAQ+LGL
Sbjct 308 LGQQEVEVDGLIAYIAGTKTLGGSIAPMDVCVRHFLQATGCSVESALEAASLHPAQMLGL 367
> hsa:51005 AMDHD2; amidohydrolase domain containing 2 (EC:3.5.1.25);
K01443 N-acetylglucosamine-6-phosphate deacetylase [EC:3.5.1.25]
Length=594
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 57/90 (63%), Positives = 58/90 (64%), Gaps = 30/90 (33%)
Query 3 LGQQEVEVDGLTAYVAG------------------------------TNTLSGSIAPMDT 32
LGQQEVEVDGLTAYVAG T TLSGSIAPMD
Sbjct 308 LGQQEVEVDGLTAYVAGERPDPLGPRSQPACQVAHDPPRACPLCSQGTKTLSGSIAPMDV 367
Query 33 CVRHFLQATGCSVESALEAASLHPAQLLGL 62
CVRHFLQATGCS+ESALEAASLHPAQLLGL
Sbjct 368 CVRHFLQATGCSMESALEAASLHPAQLLGL 397
> dre:402981 MGC77775, amdhd2; zgc:77775 (EC:3.5.1.25); K01443
N-acetylglucosamine-6-phosphate deacetylase [EC:3.5.1.25]
Length=404
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 45/60 (75%), Positives = 54/60 (90%), Gaps = 0/60 (0%)
Query 3 LGQQEVEVDGLTAYVAGTNTLSGSIAPMDTCVRHFLQATGCSVESALEAASLHPAQLLGL 62
LGQQ++++ GL AYVAGT TLSGSIA MD CVRHF +A+GC+VE+ALEAASLHPAQLLG+
Sbjct 308 LGQQQIDIQGLHAYVAGTTTLSGSIATMDMCVRHFREASGCTVEAALEAASLHPAQLLGI 367
> cel:F59B2.3 hypothetical protein; K01443 N-acetylglucosamine-6-phosphate
deacetylase [EC:3.5.1.25]
Length=418
Score = 68.9 bits (167), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 31/61 (50%), Positives = 44/61 (72%), Gaps = 0/61 (0%)
Query 2 RLGQQEVEVDGLTAYVAGTNTLSGSIAPMDTCVRHFLQATGCSVESALEAASLHPAQLLG 61
+LG Q + V GL A + GTNT +GS+A M C+RH ++ATGC +E AL++A+ PA LLG
Sbjct 319 KLGTQTIHVKGLEAKLDGTNTTAGSVASMPYCIRHLMKATGCPIEFALQSATHKPATLLG 378
Query 62 L 62
+
Sbjct 379 V 379
> hsa:7127 TNFAIP2, B94, EXOC3L3; tumor necrosis factor, alpha-induced
protein 2
Length=654
Score = 31.2 bits (69), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 14 TAYVAGTNTLSGSIAPMDTCVRHFLQATGCSVESALEAASLH 55
T + A TL IA +DT + F + GC E +EA LH
Sbjct 483 TRWAAPVETLENIIATVDTRLPEFSELQGCFREELMEALHLH 524
> eco:b0677 nagA, ECK0665, JW0663; N-acetylglucosamine-6-phosphate
deacetylase (EC:3.5.1.25); K01443 N-acetylglucosamine-6-phosphate
deacetylase [EC:3.5.1.25]
Length=382
Score = 30.8 bits (68), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 22 TLSGSIAPMDTCVRHFLQATGCSVESALEAASLHPAQLLGL 62
TLSGS M VR+ ++ G +++ L A+L+PA+ +G+
Sbjct 305 TLSGSSLTMIEGVRNLVEHCGIALDEVLRMATLYPARAIGV 345
Lambda K H
0.316 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2060478756
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40