bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4484_orf1
Length=52
Score E
Sequences producing significant alignments: (Bits) Value
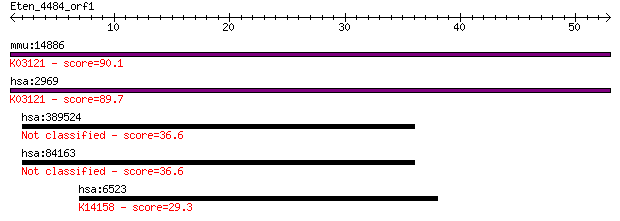
mmu:14886 Gtf2i, 6030441I21Rik, BAP-135, GtfII-I, Spin, TFII-I... 90.1 2e-18
hsa:2969 GTF2I, BAP135, BTKAP1, DIWS, FLJ38776, FLJ56355, GTFI... 89.7 2e-18
hsa:389524 GTF2IRD2B; GTF2I repeat domain containing 2B 36.6 0.019
hsa:84163 GTF2IRD2, FLJ21423, FLJ37938, FP630, GTF2IRD2A, MGC7... 36.6 0.020
hsa:6523 SLC5A1, D22S675, NAGT, SGLT1; solute carrier family 5... 29.3 2.8
> mmu:14886 Gtf2i, 6030441I21Rik, BAP-135, GtfII-I, Spin, TFII-I,
WBSCR6; general transcription factor II I; K03121 transcription
initiation factor TFII-I
Length=998
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 46/52 (88%), Positives = 49/52 (94%), Gaps = 1/52 (1%)
Query 1 AAVTLMEESEDPDYYQYNIQAGPSETDDVDDKLPLSKSLQGSHHSSEGNEGT 52
AAVT+ EESEDPDYYQYNIQ GPSETD VD+KLPLSK+LQGSHHSSEGNEGT
Sbjct 235 AAVTVKEESEDPDYYQYNIQ-GPSETDGVDEKLPLSKALQGSHHSSEGNEGT 285
> hsa:2969 GTF2I, BAP135, BTKAP1, DIWS, FLJ38776, FLJ56355, GTFII-I,
IB291, SPIN, TFII-I, WBS, WBSCR6; general transcription
factor IIi; K03121 transcription initiation factor TFII-I
Length=976
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 46/52 (88%), Positives = 48/52 (92%), Gaps = 1/52 (1%)
Query 1 AAVTLMEESEDPDYYQYNIQAGPSETDDVDDKLPLSKSLQGSHHSSEGNEGT 52
AAVT+ EESEDPDYYQYNIQ GPSETDDVD+K PLSK LQGSHHSSEGNEGT
Sbjct 235 AAVTVKEESEDPDYYQYNIQ-GPSETDDVDEKQPLSKPLQGSHHSSEGNEGT 285
> hsa:389524 GTF2IRD2B; GTF2I repeat domain containing 2B
Length=949
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 28/37 (75%), Gaps = 3/37 (8%)
Query 2 AVTLMEESEDPDYYQYNIQAG-PSETDD--VDDKLPL 35
AV++ +ESEDP+YYQYN+Q PS T + ++ +LP+
Sbjct 231 AVSVKKESEDPNYYQYNMQGSHPSSTSNEVIEMELPM 267
> hsa:84163 GTF2IRD2, FLJ21423, FLJ37938, FP630, GTF2IRD2A, MGC75203;
GTF2I repeat domain containing 2
Length=949
Score = 36.6 bits (83), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 28/37 (75%), Gaps = 3/37 (8%)
Query 2 AVTLMEESEDPDYYQYNIQAG-PSETDD--VDDKLPL 35
AV++ +ESEDP+YYQYN+Q PS T + ++ +LP+
Sbjct 231 AVSVKKESEDPNYYQYNMQGSHPSSTSNEVIEMELPM 267
> hsa:6523 SLC5A1, D22S675, NAGT, SGLT1; solute carrier family
5 (sodium/glucose cotransporter), member 1; K14158 solute carrier
family 5 (sodium/glucose cotransporter), member 1
Length=664
Score = 29.3 bits (64), Expect = 2.8, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 7 EESEDPDYYQYNIQAGPSETDDVDDKLPLSK 37
EE D D + NIQ GP ET +++ ++P K
Sbjct 568 EERIDLDAEEENIQEGPKETIEIETQVPEKK 598
Lambda K H
0.299 0.122 0.335
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022656792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40