bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4472_orf2
Length=57
Score E
Sequences producing significant alignments: (Bits) Value
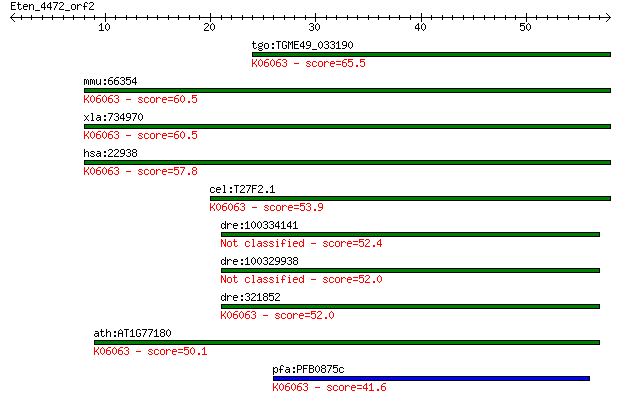
tgo:TGME49_033190 hypothetical protein ; K06063 SNW domain-con... 65.5 4e-11
mmu:66354 Snw1, 2310008B08Rik, AW048543, MGC55033, NCoA-62, SK... 60.5 1e-09
xla:734970 snw1, MGC132028, bx42, ncoa-62, prp45, prpf45, skii... 60.5 1e-09
hsa:22938 SNW1, Bx42, MGC119379, NCOA-62, PRPF45, Prp45, SKIIP... 57.8 8e-09
cel:T27F2.1 skp-1; mammalian SKIP (Ski interacting protein) ho... 53.9 1e-07
dre:100334141 SKI-interacting protein-like 52.4 4e-07
dre:100329938 SKI-interacting protein-like 52.0 4e-07
dre:321852 snw1, MGC123090, fb37b08, skiip, wu:fb37b08, zgc:12... 52.0 5e-07
ath:AT1G77180 chromatin protein family; K06063 SNW domain-cont... 50.1 2e-06
pfa:PFB0875c chromatin-binding protein, putative; K06063 SNW d... 41.6 6e-04
> tgo:TGME49_033190 hypothetical protein ; K06063 SNW domain-containing
protein 1
Length=557
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 27/34 (79%), Positives = 30/34 (88%), Gaps = 0/34 (0%)
Query 24 PPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPLG 57
PPY R+GF+PR EEDFGGGGA+PEI VAQFPLG
Sbjct 83 PPYGQRKGFVPRSEEDFGGGGAFPEILVAQFPLG 116
> mmu:66354 Snw1, 2310008B08Rik, AW048543, MGC55033, NCoA-62,
SKIP, Skiip; SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 29/50 (58%), Positives = 36/50 (72%), Gaps = 0/50 (0%)
Query 8 AQSRLQKSQKAPAREIPPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPLG 57
+Q LQ S + RE PPY +R+G+IPR EDFG GGA+PEI VAQ+PL
Sbjct 26 SQRSLQTSLVSSRREPPPYGYRKGWIPRLLEDFGDGGAFPEIHVAQYPLD 75
> xla:734970 snw1, MGC132028, bx42, ncoa-62, prp45, prpf45, skiip;
SNW domain containing 1; K06063 SNW domain-containing protein
1
Length=535
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 28/50 (56%), Positives = 35/50 (70%), Gaps = 0/50 (0%)
Query 8 AQSRLQKSQKAPAREIPPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPLG 57
AQ Q + + RE PPY HR+G++PR EDFG GGA+PEI VAQ+PL
Sbjct 26 AQKSRQTALVSSRREPPPYGHRKGWVPRSLEDFGDGGAFPEIHVAQYPLD 75
> hsa:22938 SNW1, Bx42, MGC119379, NCOA-62, PRPF45, Prp45, SKIIP,
SKIP; SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 28/50 (56%), Positives = 35/50 (70%), Gaps = 0/50 (0%)
Query 8 AQSRLQKSQKAPAREIPPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPLG 57
+Q Q S + RE PPY +R+G+IPR EDFG GGA+PEI VAQ+PL
Sbjct 26 SQRSRQTSLVSSRREPPPYGYRKGWIPRLLEDFGDGGAFPEIHVAQYPLD 75
> cel:T27F2.1 skp-1; mammalian SKIP (Ski interacting protein)
homolog family member (skp-1); K06063 SNW domain-containing
protein 1
Length=535
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 25/38 (65%), Positives = 28/38 (73%), Gaps = 0/38 (0%)
Query 20 AREIPPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPLG 57
++E PPY R F PR EDFG GGA+PEI VAQFPLG
Sbjct 41 SKEPPPYGKRTSFRPRGPEDFGDGGAFPEIHVAQFPLG 78
> dre:100334141 SKI-interacting protein-like
Length=344
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/36 (61%), Positives = 28/36 (77%), Gaps = 0/36 (0%)
Query 21 REIPPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPL 56
R+ PPY R+ ++PR EDFG GGA+PEI VAQ+PL
Sbjct 39 RDPPPYGFRKSWVPRALEDFGDGGAFPEIHVAQYPL 74
> dre:100329938 SKI-interacting protein-like
Length=464
Score = 52.0 bits (123), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 22/36 (61%), Positives = 28/36 (77%), Gaps = 0/36 (0%)
Query 21 REIPPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPL 56
R+ PPY R+ ++PR EDFG GGA+PEI VAQ+PL
Sbjct 39 RDPPPYGFRKSWVPRALEDFGDGGAFPEIHVAQYPL 74
> dre:321852 snw1, MGC123090, fb37b08, skiip, wu:fb37b08, zgc:123090;
SNW domain containing 1; K06063 SNW domain-containing
protein 1
Length=536
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/36 (61%), Positives = 28/36 (77%), Gaps = 0/36 (0%)
Query 21 REIPPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPL 56
R+ PPY R+ ++PR EDFG GGA+PEI VAQ+PL
Sbjct 39 RDPPPYGFRKSWVPRALEDFGDGGAFPEIHVAQYPL 74
> ath:AT1G77180 chromatin protein family; K06063 SNW domain-containing
protein 1
Length=613
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 23/48 (47%), Positives = 31/48 (64%), Gaps = 0/48 (0%)
Query 9 QSRLQKSQKAPAREIPPYLHRQGFIPRCEEDFGGGGAYPEIFVAQFPL 56
+S KS + +P YL+RQG P+ EDFG GGA+PEI + Q+PL
Sbjct 32 ESETVKSSSIKFKVVPAYLNRQGLRPKNPEDFGDGGAFPEIHLPQYPL 79
> pfa:PFB0875c chromatin-binding protein, putative; K06063 SNW
domain-containing protein 1
Length=482
Score = 41.6 bits (96), Expect = 6e-04, Method: Composition-based stats.
Identities = 17/30 (56%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 26 YLHRQGFIPRCEEDFGGGGAYPEIFVAQFP 55
YL R+ C EDF GGGAYPEI + Q+P
Sbjct 47 YLKRRHLRITCNEDFQGGGAYPEIHMNQYP 76
Lambda K H
0.321 0.141 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2082685716
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40