bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4403_orf1
Length=194
Score E
Sequences producing significant alignments: (Bits) Value
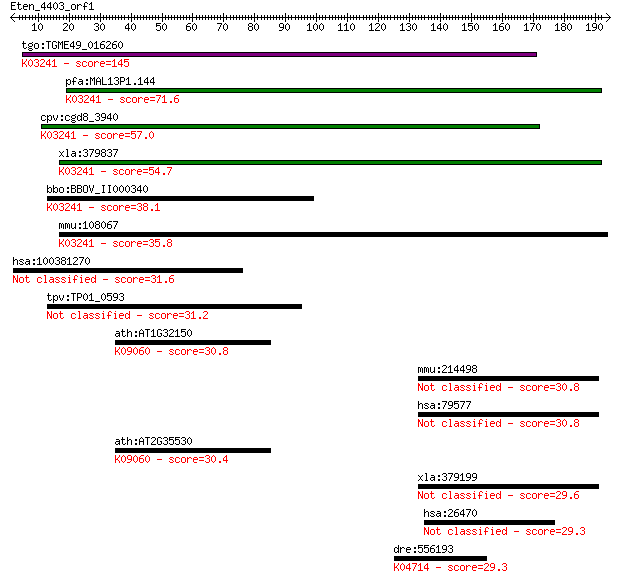
tgo:TGME49_016260 eukaryotic initiation factor-2B gamma subuni... 145 6e-35
pfa:MAL13P1.144 translation initiation factor EIF-2B gamma sub... 71.6 1e-12
cpv:cgd8_3940 eIF-2B gamma, eukaryotic translation initiation ... 57.0 4e-08
xla:379837 eif2b3, MGC52809; eukaryotic translation initiation... 54.7 2e-07
bbo:BBOV_II000340 18.m06011; hypothetical protein; K03241 tran... 38.1 0.020
mmu:108067 Eif2b3, 1190002P15Rik; eukaryotic translation initi... 35.8 0.094
hsa:100381270 ZBED6, MGR; zinc finger, BED-type containing 6 31.6
tpv:TP01_0593 hypothetical protein 31.2 2.0
ath:AT1G32150 bZIP transcription factor family protein; K09060... 30.8 2.8
mmu:214498 Cdc73, 8430414L16Rik, BC027756, C130030P16Rik, C812... 30.8 2.8
hsa:79577 CDC73, C1orf28, FLJ23316, HPTJT, HRPT2, HYX; cell di... 30.8 2.8
ath:AT2G35530 bZIP transcription factor family protein; K09060... 30.4 3.7
xla:379199 cdc73, MGC53995; cell division cycle 73, Paf1/RNA p... 29.6 7.0
hsa:26470 SEZ6L2, FLJ90517, PSK-1; seizure related 6 homolog (... 29.3 7.8
dre:556193 sgms2; sphingomyelin synthase 2 (EC:2.7.8.27); K047... 29.3 9.8
> tgo:TGME49_016260 eukaryotic initiation factor-2B gamma subunit,
putative (EC:2.7.7.27 2.7.7.60); K03241 translation initiation
factor eIF-2B subunit gamma
Length=492
Score = 145 bits (367), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 74/171 (43%), Positives = 103/171 (60%), Gaps = 22/171 (12%)
Query 5 SDTSRGNPVTVVVDDAGVQLLAIQDRHSMSHGSSLQIPKLHLFLYPSQHLLPHYYDPHVY 64
D ++GNPV + ++ LL IQDRHS+SHG+ L IPKL LF +PS + + YDPHVY
Sbjct 219 KDANKGNPVAIAFEETETLLLGIQDRHSISHGAQLAIPKLTLFYHPSVFVKANLYDPHVY 278
Query 65 VFTHATLKIFDQPSLRHSLYSIRFDLVPYMTTMQMTSAAEVWSNSRLECDIFEQKLWAI- 123
+F + LKI + P LR++L SIRFDLVPYM+ MQMT A +WS+SRL+CD+F++ L +
Sbjct 279 LFKLSALKILEDPKLRNTLTSIRFDLVPYMSLMQMTPQASLWSSSRLDCDVFDELLDSFD 338
Query 124 ----QHQGEEESTSTSQTATAPTNEMPTGSGLPVHYTAANPPPRPAKGNRV 170
+ + +E+ S Q YT AN P +P NR+
Sbjct 339 EPHKKREDKEQDRSREQ-----------------GYTLANRPEQPFCSNRM 372
> pfa:MAL13P1.144 translation initiation factor EIF-2B gamma subunit,
putative; K03241 translation initiation factor eIF-2B
subunit gamma
Length=473
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/173 (24%), Positives = 83/173 (47%), Gaps = 22/173 (12%)
Query 19 DAGVQLLAIQDRHSMSHGSSLQIPKLHLFLYPSQHLLPHYYDPHVYVFTHATLKIFDQPS 78
D ++++I+D SM ++I K++L + + L D HVY+F H L+I +Q
Sbjct 194 DKNSKVVSIKDSLSMKENGKMKISKVNLLFHKNFVLKTDLLDSHVYIFKHYVLEIMEQKK 253
Query 79 LRHSLYSIRFDLVPYMTTMQMTSAAEVWSNSRLECDIFEQKLWAIQHQGEEESTSTSQTA 138
+ S SI++DL+PY+ +Q TS A +S + +++ + +++G++
Sbjct 254 NKFS--SIKYDLIPYLVKIQNTSKAAEYSKGEFKYNMYNTLIE--KYEGDD--------- 300
Query 139 TAPTNEMPTGSGLPVHYTAANPPPRPAKGNRVVCCIHPEAAGICCRVNNLADY 191
E+ G + N + VVC I P+ G C R+N++ ++
Sbjct 301 -----EIEEGKRENLMLDIINNENVES----VVCYIQPKNNGFCQRINSIPNF 344
> cpv:cgd8_3940 eIF-2B gamma, eukaryotic translation initiation
factor 2B subunit 3 that has a nucleotide diphospho sugar
transferase at the N-terminus and a UDP N-acetylglucosamine
acyltransferase at the C-terminus ; K03241 translation initiation
factor eIF-2B subunit gamma
Length=500
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 41/174 (23%), Positives = 77/174 (44%), Gaps = 23/174 (13%)
Query 11 NPVTVVVDDAGVQLLAIQDRHSMSHGSSL-QIPKLHLFLYPSQHLLPHYYDPHVYVFTHA 69
N V+D+ +L+I+D +S + + ++ KL LF +P+ L D HVY+F +
Sbjct 206 NRSIFVIDEKDEVILSIKDFYSAKQENEVSELSKLQLFWHPNVSLRTDLVDLHVYLFKSS 265
Query 70 TLKIFDQPSLRH----------SLYSIRFDLVPYMTTMQMTSAAEVWSNSRLECDIFEQK 119
KI + S + SIR +L+P++ Q +E+W S+ +C
Sbjct 266 IFKILEIASGSQKISTIEYPEDGIESIRLELLPFLAKNQHVPGSELWGRSKFDC------ 319
Query 120 LWAIQHQGEEESTSTSQTATAPT--NEMPTGSGLPVHYTAANPPPRPAKGNRVV 171
H ++E T+++ ++ T + P G V Y P ++ N ++
Sbjct 320 ----YHFLDDEITTSNDSSVKFTKIDLPPKIEGTSVSYFLQKLPQNSSRVNTIM 369
> xla:379837 eif2b3, MGC52809; eukaryotic translation initiation
factor 2B, subunit 3 gamma, 58kDa; K03241 translation initiation
factor eIF-2B subunit gamma
Length=456
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 48/176 (27%), Positives = 75/176 (42%), Gaps = 16/176 (9%)
Query 17 VDDAGVQLLAIQDRHSMSHGSSLQIPKLHLFLYPSQHLLPHYYDPHVYVFTHATLKIFDQ 76
VDD G +LL + + + G L + + L YP H+ D H+Y I D
Sbjct 163 VDDKGTRLLLLANEEDLDDG--LNLKRSLLQRYPRIHIKMGMVDAHLYCLRKY---IVDF 217
Query 77 PSLRHSLYSIRFDLVPYMTTMQMTSAAEVWSNSRLECDIFEQKLWAIQHQGEEESTSTSQ 136
S SIR +L+PY+ Q S SNS+ ++K +H G +ES
Sbjct 218 LHTNESFSSIRRELIPYLVRKQFLSV----SNSQ------QKKEEQEEHNGGKESLPGDI 267
Query 137 TATAPTNEMPTGSGLPVHYTAANPPPR-PAKGNRVVCCIHPEAAGICCRVNNLADY 191
+ +++ + + R P G+R+ C +H +CCRVN+LA Y
Sbjct 268 YSFITQDKLLDRALEMSCWNDHRGDMREPYHGSRLRCYVHVAGNELCCRVNSLAMY 323
> bbo:BBOV_II000340 18.m06011; hypothetical protein; K03241 translation
initiation factor eIF-2B subunit gamma
Length=398
Score = 38.1 bits (87), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 41/86 (47%), Gaps = 2/86 (2%)
Query 13 VTVVVDDAGVQLLAIQDRHSMSHGSSLQIPKLHLFLYPSQHLLPHYYDPHVYVFTHATLK 72
V ++D ++++I + S+ G I K + + YD H+YVF+ +
Sbjct 196 VLAMLDIDHSKVVSISNYLSLCSGEPTNISKWTFRNHNKCSIRCDLYDAHIYVFSKDIIH 255
Query 73 IFDQPSLRHSLYSIRFDLVPYMTTMQ 98
+ + + S S+R D++PY+ MQ
Sbjct 256 MLTEKCFKQS--SLRLDVIPYIIAMQ 279
> mmu:108067 Eif2b3, 1190002P15Rik; eukaryotic translation initiation
factor 2B, subunit 3; K03241 translation initiation
factor eIF-2B subunit gamma
Length=452
Score = 35.8 bits (81), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 43/182 (23%), Positives = 69/182 (37%), Gaps = 25/182 (13%)
Query 17 VDDAGVQLLAIQDRHSMSHGSSLQIPKLHLFLYPSQHLLPHYYDPHVYVFTHATLKIFDQ 76
VD G +LL + + + L I L +P H D H+Y + +
Sbjct 160 VDSTGKRLLFMANEADLDE--ELVIKGSILQKHPRIHFHTGLVDAHLYCLKKYVVDFLME 217
Query 77 PSLRHSLYSIRFDLVPYMTTMQMTSAAEVWSNSRLECDIFEQKLWAIQHQGEEESTSTSQ 136
S+ SIR +L+PY+ Q +SA+ E D+ +++L ++ + T
Sbjct 218 ---NRSITSIRSELIPYLVRKQFSSASSQQRQEDKEEDLKKKELKSLDIYSFIKKDDT-- 272
Query 137 TATAPTNEMPTGSGLPVHYTAANPPPRPAK-----GNRVVCCIHPEAAGICCRVNNLADY 191
AP Y A R K ++V C +H G+C RV+ L Y
Sbjct 273 LTLAP-------------YDACWNAFRGDKWEDLSRSQVRCYVHIMKEGLCSRVSTLGLY 319
Query 192 RE 193
E
Sbjct 320 ME 321
> hsa:100381270 ZBED6, MGR; zinc finger, BED-type containing 6
Length=979
Score = 31.6 bits (70), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 39/75 (52%), Gaps = 8/75 (10%)
Query 2 QEKSDTSRGNPVTVVVDDAGVQLLAIQDRHSMSHGSSLQIPKLHLFLYPSQHLLPHYYDP 61
QE+ T GNPV+ + A +Q++ ++D H ++ S+ P F+ Q + P Y P
Sbjct 395 QERETTCCGNPVSSHISQAIIQMI-VEDMHPYNYFST---PAFQRFM---QIVAPDYRLP 447
Query 62 -HVYVFTHATLKIFD 75
Y FT A +++D
Sbjct 448 SETYFFTKAVPQLYD 462
> tpv:TP01_0593 hypothetical protein
Length=382
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 39/88 (44%), Gaps = 6/88 (6%)
Query 13 VTVVVDDAGVQLLAIQDRHSMSHGSSLQIPKLHLFLYPSQHLLPHYYDPHVYVFTHATLK 72
V +D + LL+I S+ G S Q+ + HL + + + D HVY F+ K
Sbjct 190 VVTALDKSTCSLLSIAPHLSVESGESFQLFRYHLVNHANSLITFDLVDIHVYSFSINIFK 249
Query 73 IFDQPSL------RHSLYSIRFDLVPYM 94
+ L R+++Y+I V Y+
Sbjct 250 VLRCDFLLNSSIRRYTIYTIVQHTVEYL 277
> ath:AT1G32150 bZIP transcription factor family protein; K09060
plant G-box-binding factor
Length=389
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 1/51 (1%)
Query 35 HGSSLQIPKLHLFLYPSQHLLPHYYD-PHVYVFTHATLKIFDQPSLRHSLY 84
HG P+ H +++ QH++P Y PH YV + ++ PSL Y
Sbjct 61 HGYVASSPQPHPYMWGVQHMMPPYGTPPHPYVTMYPPGGMYAHPSLPPGSY 111
> mmu:214498 Cdc73, 8430414L16Rik, BC027756, C130030P16Rik, C81219,
Hrpt2, MGC29274, MGC36559; cell division cycle 73, Paf1/RNA
polymerase II complex component, homolog (S. cerevisiae)
Length=531
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 25/58 (43%), Gaps = 0/58 (0%)
Query 133 STSQTATAPTNEMPTGSGLPVHYTAANPPPRPAKGNRVVCCIHPEAAGICCRVNNLAD 190
S ++ A+A + P +P + A PPP KG+R I P A + N D
Sbjct 322 SVTEGASARKTQTPAAQPVPRPVSQARPPPNQKKGSRTPIIIIPAATTSLITMLNAKD 379
> hsa:79577 CDC73, C1orf28, FLJ23316, HPTJT, HRPT2, HYX; cell
division cycle 73, Paf1/RNA polymerase II complex component,
homolog (S. cerevisiae)
Length=531
Score = 30.8 bits (68), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 25/58 (43%), Gaps = 0/58 (0%)
Query 133 STSQTATAPTNEMPTGSGLPVHYTAANPPPRPAKGNRVVCCIHPEAAGICCRVNNLAD 190
S ++ A+A + P +P + A PPP KG+R I P A + N D
Sbjct 322 SVTEGASARKTQTPAAQPVPRPVSQARPPPNQKKGSRTPIIIIPAATTSLITMLNAKD 379
> ath:AT2G35530 bZIP transcription factor family protein; K09060
plant G-box-binding factor
Length=409
Score = 30.4 bits (67), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 25/51 (49%), Gaps = 1/51 (1%)
Query 35 HGSSLQIPKLHLFLYPSQHLLPHYYD-PHVYVFTHATLKIFDQPSLRHSLY 84
HG P+ H +++ QH++P Y PH YV + ++ PS+ Y
Sbjct 57 HGYVASSPQPHPYMWGVQHMMPPYGTPPHPYVAMYPPGGMYAHPSMPPGSY 107
> xla:379199 cdc73, MGC53995; cell division cycle 73, Paf1/RNA
polymerase II complex component, homolog
Length=531
Score = 29.6 bits (65), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 17/58 (29%), Positives = 25/58 (43%), Gaps = 0/58 (0%)
Query 133 STSQTATAPTNEMPTGSGLPVHYTAANPPPRPAKGNRVVCCIHPEAAGICCRVNNLAD 190
S ++ A+A + P +P + A PPP KG+R I P A + N D
Sbjct 322 SVTEGASARKTQTPAVQPVPRPVSQARPPPNQKKGSRTPIIIIPAATTSLITMLNAKD 379
> hsa:26470 SEZ6L2, FLJ90517, PSK-1; seizure related 6 homolog
(mouse)-like 2
Length=840
Score = 29.3 bits (64), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 24/55 (43%), Gaps = 13/55 (23%)
Query 135 SQTATAPTNEM---------PTGSGLPVHYTA----ANPPPRPAKGNRVVCCIHP 176
Q +PTN + P G G +HY A PPRPA G+ V +HP
Sbjct 184 GQVLRSPTNRLLLHFQSPRVPRGGGFRIHYQAYLLSCGFPPRPAHGDVSVTDLHP 238
> dre:556193 sgms2; sphingomyelin synthase 2 (EC:2.7.8.27); K04714
shingomyelin synthase [EC:2.7.8.27]
Length=351
Score = 29.3 bits (64), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 20/30 (66%), Gaps = 4/30 (13%)
Query 125 HQGEEESTSTSQTATAPTNEMPTGSGLPVH 154
HQG +E T+TS+T AP+ TG+ PVH
Sbjct 6 HQGNKEPTNTSRTDGAPS----TGATCPVH 31
Lambda K H
0.318 0.131 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5608917492
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40