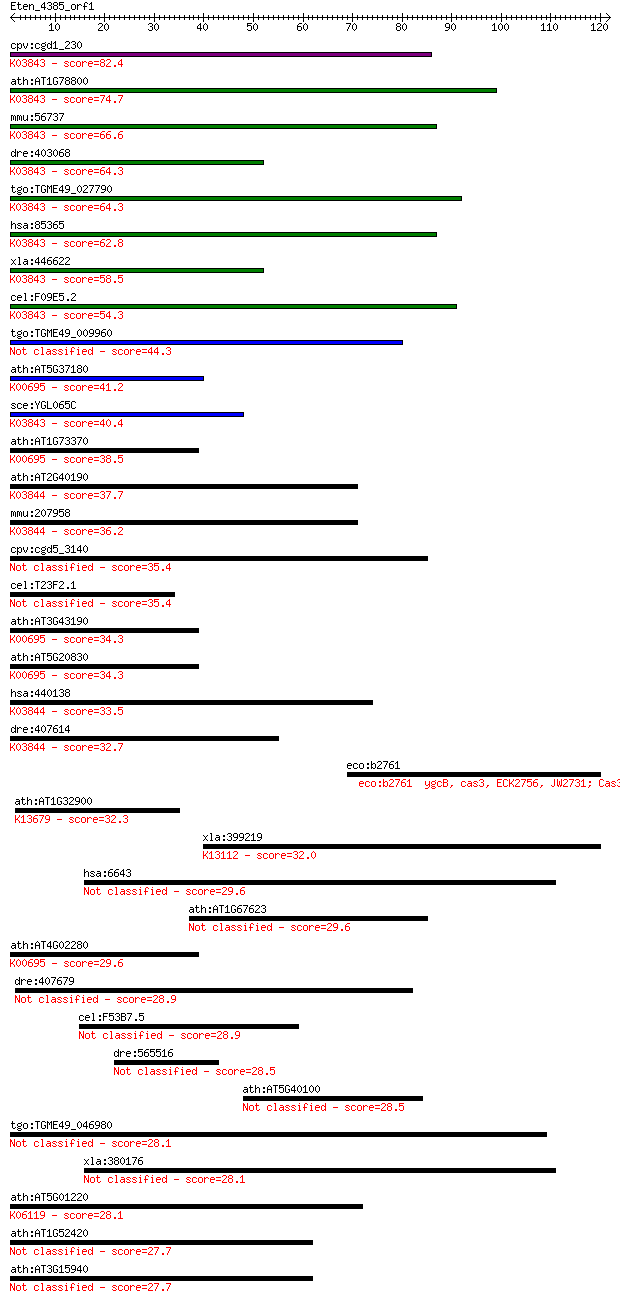
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4385_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
cpv:cgd1_230 ALG-2 like alpha-1,3 mannosyltransferase ; K03843... 82.4 3e-16
ath:AT1G78800 glycosyl transferase family 1 protein; K03843 al... 74.7 7e-14
mmu:56737 Alg2, 1110018A23Rik, 1300013N08Rik, ALPG2, CDGIi, MN... 66.6 2e-11
dre:403068 alg2, FLJ14511, im:7145131; asparagine-linked glyco... 64.3 9e-11
tgo:TGME49_027790 glycosyl transferase, group 1 domain contain... 64.3 1e-10
hsa:85365 ALG2, CDGIi, FLJ14511, NET38, hALPG2; asparagine-lin... 62.8 2e-10
xla:446622 alg2, MGC82301; asparagine-linked glycosylation 2 h... 58.5 4e-09
cel:F09E5.2 hypothetical protein; K03843 alpha-1,3/alpha-1,6-m... 54.3 1e-07
tgo:TGME49_009960 glycan synthetase, putative (EC:2.4.1.21 2.4... 44.3 8e-05
ath:AT5G37180 SUS5; SUS5; UDP-glycosyltransferase/ sucrose syn... 41.2 0.001
sce:YGL065C ALG2; Alg2p (EC:2.4.1.132 2.4.1.-); K03843 alpha-1... 40.4 0.001
ath:AT1G73370 SUS6; SUS6 (SUCROSE SYNTHASE 6); UDP-glycosyltra... 38.5 0.005
ath:AT2G40190 glycosyl transferase family 1 protein; K03844 al... 37.7 0.010
mmu:207958 Alg11, AI849156, AW492253, B230397C21; asparagine-l... 36.2 0.028
cpv:cgd5_3140 LPS glycosyltransferase of possible cyanobacteri... 35.4 0.041
cel:T23F2.1 bus-8; Bacterially Un-Swollen (M. nematophilum res... 35.4 0.043
ath:AT3G43190 SUS4; SUS4; UDP-glycosyltransferase/ sucrose syn... 34.3 0.087
ath:AT5G20830 SUS1; SUS1 (SUCROSE SYNTHASE 1); UDP-glycosyltra... 34.3 0.11
hsa:440138 ALG11, CDG1P, GT8, KIAA0266, UTP14C; asparagine-lin... 33.5 0.18
dre:407614 alg11; asparagine-linked glycosylation 11, alpha-1,... 32.7 0.25
eco:b2761 ygcB, cas3, ECK2756, JW2731; Cas3 predicted helicase... 32.7 0.28
ath:AT1G32900 starch synthase, putative; K13679 granule-bound ... 32.3 0.39
xla:399219 thrap3, MGC132228, trap150; thyroid hormone recepto... 32.0 0.48
hsa:6643 SNX2, MGC5204, TRG-9; sorting nexin 2 29.6
ath:AT1G67623 F-box family protein 29.6 2.3
ath:AT4G02280 SUS3; SUS3 (sucrose synthase 3); UDP-glycosyltra... 29.6 2.3
dre:407679 topbp1; topoisomerase (DNA) II binding protein 1 28.9
cel:F53B7.5 hypothetical protein 28.9 3.8
dre:565516 hypothetical LOC565516 28.5 5.7
ath:AT5G40100 disease resistance protein (TIR-NBS-LRR class), ... 28.5 6.0
tgo:TGME49_046980 hypothetical protein 28.1 6.7
xla:380176 snx2, MGC64590; sorting nexin 2 28.1
ath:AT5G01220 SQD2; SQD2 (sulfoquinovosyldiacylglycerol 2); UD... 28.1 7.7
ath:AT1G52420 glycosyl transferase family 1 protein 27.7 8.5
ath:AT3G15940 glycosyl transferase family 1 protein 27.7 9.1
> cpv:cgd1_230 ALG-2 like alpha-1,3 mannosyltransferase ; K03843
alpha-1,3/alpha-1,6-mannosyltransferase [EC:2.4.1.132 2.4.1.-]
Length=474
Score = 82.4 bits (202), Expect = 3e-16, Method: Composition-based stats.
Identities = 42/85 (49%), Positives = 55/85 (64%), Gaps = 1/85 (1%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FG+VPCEAM +G +ACNSGGP ETIV G+TGFLCEP P +F+ + ++ +SRDS E
Sbjct 371 FGMVPCEAMSVGTCVIACNSGGPTETIVHGKTGFLCEPNPENFAIRMNELIKISRDSIES 430
Query 61 LAALRSKAEAHAACCFSPDVFRRSL 85
S E A FS +F++ L
Sbjct 431 SVWSSSCKERVTA-LFSQSIFQKRL 454
> ath:AT1G78800 glycosyl transferase family 1 protein; K03843
alpha-1,3/alpha-1,6-mannosyltransferase [EC:2.4.1.132 2.4.1.-]
Length=403
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 40/98 (40%), Positives = 55/98 (56%), Gaps = 4/98 (4%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FGIVP EAM +ACNSGGP ET+ +G TG+LCEPTP FS+A+ R + ++PE
Sbjct 309 FGIVPLEAMAAYKPVIACNSGGPVETVKNGVTGYLCEPTPEDFSSAMARFI----ENPEL 364
Query 61 LAALRSKAEAHAACCFSPDVFRRSLHGILEEAAGPSDE 98
+ ++A H FS F + L+ L + E
Sbjct 365 ANRMGAEARNHVVESFSVKTFGQKLNQYLVDVVSSPKE 402
> mmu:56737 Alg2, 1110018A23Rik, 1300013N08Rik, ALPG2, CDGIi,
MNCb-5081; asparagine-linked glycosylation 2 homolog (yeast,
alpha-1,3-mannosyltransferase); K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=415
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 36/86 (41%), Positives = 48/86 (55%), Gaps = 4/86 (4%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FGIVP EAM++ +A N+GGP E+IV TGFLCEP P FS A+++ + P
Sbjct 327 FGIVPLEAMYMQCPVIAVNNGGPLESIVHKVTGFLCEPDPVHFSEAMEKFI----HKPSL 382
Query 61 LAALRSKAEAHAACCFSPDVFRRSLH 86
A + +A A FS D F L+
Sbjct 383 KATMGLAGKARVAEKFSADAFADQLY 408
> dre:403068 alg2, FLJ14511, im:7145131; asparagine-linked glycosylation
2 homolog (S. cerevisiae, alpha-1,3-mannosyltransferase);
K03843 alpha-1,3/alpha-1,6-mannosyltransferase [EC:2.4.1.132
2.4.1.-]
Length=402
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 28/51 (54%), Positives = 36/51 (70%), Gaps = 0/51 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRIL 51
FGIVP E+M++ +A NSGGP E++ ETGFLCEPTP FS A+Q +
Sbjct 313 FGIVPIESMYLRCPVIAVNSGGPLESVAHEETGFLCEPTPERFSEAMQNFV 363
> tgo:TGME49_027790 glycosyl transferase, group 1 domain containing
protein (EC:2.4.1.14); K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=506
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 48/91 (52%), Gaps = 0/91 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FG+VP EA +G VA NSGGPRE+I+ G+TGFLCE SF ++ ++ + R P
Sbjct 403 FGMVPLEANALGCPVVASNSGGPRESILHGKTGFLCEHDAGSFGDSILMLVRMQRGEPHI 462
Query 61 LAALRSKAEAHAACCFSPDVFRRSLHGILEE 91
+R A H F F L ++ E
Sbjct 463 YTEMRENARRHVKENFDCGAFGARLRQLVAE 493
> hsa:85365 ALG2, CDGIi, FLJ14511, NET38, hALPG2; asparagine-linked
glycosylation 2, alpha-1,3-mannosyltransferase homolog
(S. cerevisiae) (EC:2.4.1.-); K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=416
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/86 (40%), Positives = 46/86 (53%), Gaps = 4/86 (4%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FGIVP EAM++ +A NSGGP E+I TGFLCEP P FS A+++ + P
Sbjct 327 FGIVPLEAMYMQCPVIAVNSGGPLESIDHSVTGFLCEPDPVHFSEAIEKFI----REPSL 382
Query 61 LAALRSKAEAHAACCFSPDVFRRSLH 86
A + A FSP+ F L+
Sbjct 383 KATMGLAGRARVKEKFSPEAFTEQLY 408
> xla:446622 alg2, MGC82301; asparagine-linked glycosylation 2
homolog; K03843 alpha-1,3/alpha-1,6-mannosyltransferase [EC:2.4.1.132
2.4.1.-]
Length=404
Score = 58.5 bits (140), Expect = 4e-09, Method: Composition-based stats.
Identities = 25/51 (49%), Positives = 35/51 (68%), Gaps = 0/51 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRIL 51
FGIVP EAM++ VA NSGGP E++ + TGFLC P P F+ A+++ +
Sbjct 315 FGIVPIEAMYMHCPVVAVNSGGPLESVENNVTGFLCSPNPKEFADAMEKFV 365
> cel:F09E5.2 hypothetical protein; K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=400
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 30/90 (33%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FGIVP EAM++G +A N+GGP E++ + ETGFL + T +F+ ++++ L +D E
Sbjct 312 FGIVPVEAMYLGTPVIAVNTGGPCESVRNNETGFLVDQTAEAFA---EKMIDLMKDE-EM 367
Query 61 LAALRSKAEAHAACCFSPDVFRRSLHGILE 90
+ + F+ + F R L I++
Sbjct 368 YRRMSEEGPKWVQKVFAFEAFARKLDEIIQ 397
> tgo:TGME49_009960 glycan synthetase, putative (EC:2.4.1.21 2.4.1.18)
Length=1707
Score = 44.3 bits (103), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 38/79 (48%), Gaps = 4/79 (5%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FGIV EA G VA NSGGPR+ + TG+L +P P S + IL + E
Sbjct 1354 FGIVVLEAWSSGKPVVATNSGGPRDFVNPHHTGYLVDPEPGSIAWGCCEIL----KNFEH 1409
Query 61 LAALRSKAEAHAACCFSPD 79
+ S+ AA FS D
Sbjct 1410 SRWMGSRGRVTAAFSFSWD 1428
> ath:AT5G37180 SUS5; SUS5; UDP-glycosyltransferase/ sucrose synthase
(EC:2.4.1.13); K00695 sucrose synthase [EC:2.4.1.13]
Length=836
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 19/39 (48%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPT 39
FG+ EAM G + A N GGP E IVDG +GF +P+
Sbjct 671 FGLTVIEAMSCGLVTFATNQGGPAEIIVDGVSGFHIDPS 709
> sce:YGL065C ALG2; Alg2p (EC:2.4.1.132 2.4.1.-); K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=503
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 25/55 (45%), Positives = 32/55 (58%), Gaps = 8/55 (14%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETI---VDGE-----TGFLCEPTPASFSAAL 47
FGIVP EAM +G +A N+GGP ETI V GE TG+L P ++ A+
Sbjct 337 FGIVPLEAMKLGKPVLAVNNGGPLETIKSYVAGENESSATGWLKPAVPIQWATAI 391
> ath:AT1G73370 SUS6; SUS6 (SUCROSE SYNTHASE 6); UDP-glycosyltransferase/
sucrose synthase; K00695 sucrose synthase [EC:2.4.1.13]
Length=942
Score = 38.5 bits (88), Expect = 0.005, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEP 38
FG+ EAM G A N GGP E IVDG +GF +P
Sbjct 682 FGLTVIEAMNCGLPTFATNQGGPAEIIVDGVSGFHIDP 719
> ath:AT2G40190 glycosyl transferase family 1 protein; K03844
alpha-1,2-mannosyltransferase [EC:2.4.1.-]
Length=463
Score = 37.7 bits (86), Expect = 0.010, Method: Composition-based stats.
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 5/74 (6%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIV---DGE-TGFLCEPTPASFSAALQRILLLSRD 56
FGI E M G +P+A NS GP+ IV DG+ TGFL E T ++ A+ I+ ++
Sbjct 367 FGISVVEYMAAGAIPIAHNSAGPKMDIVLEEDGQKTGFLAE-TVEEYAEAILEIVKMNET 425
Query 57 SPEELAALRSKAEA 70
++A K A
Sbjct 426 ERLKMAESARKRAA 439
> mmu:207958 Alg11, AI849156, AW492253, B230397C21; asparagine-linked
glycosylation 11 homolog (yeast, alpha-1,2-mannosyltransferase)
(EC:2.4.-.-); K03844 alpha-1,2-mannosyltransferase
[EC:2.4.1.-]
Length=450
Score = 36.2 bits (82), Expect = 0.028, Method: Composition-based stats.
Identities = 29/74 (39%), Positives = 40/74 (54%), Gaps = 8/74 (10%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIV---DGE-TGFLCEPTPASFSAALQRILLLSRD 56
FGI E M G + +A NSGGP+ IV +G+ TGFL E + ++ ++ IL L
Sbjct 358 FGIGVVECMAAGTVILAHNSGGPKLDIVIPHEGQITGFLAE-SEEGYADSMAHILSL--- 413
Query 57 SPEELAALRSKAEA 70
S EE +R A A
Sbjct 414 SAEERLQIRKNARA 427
> cpv:cgd5_3140 LPS glycosyltransferase of possible cyanobacterial
origin
Length=2069
Score = 35.4 bits (80), Expect = 0.041, Method: Composition-based stats.
Identities = 27/84 (32%), Positives = 36/84 (42%), Gaps = 4/84 (4%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FGIV E G VA SGGPR+ + G+L EP S + IL + +
Sbjct 1548 FGIVVLEGWTAGKPVVATTSGGPRDFLTPNVDGYLVEPCKDSIAWGCCEIL----KNFDH 1603
Query 61 LAALRSKAEAHAACCFSPDVFRRS 84
+ S+ AA FS D R+
Sbjct 1604 SRWMGSRGRVKAAYSFSWDSIARA 1627
> cel:T23F2.1 bus-8; Bacterially Un-Swollen (M. nematophilum resistant)
family member (bus-8)
Length=437
Score = 35.4 bits (80), Expect = 0.043, Method: Composition-based stats.
Identities = 16/33 (48%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETG 33
FGIVP EA+ + C+SGGP ET+++ TG
Sbjct 298 FGIVPIEALDQRRPVIVCDSGGPAETVLEDITG 330
> ath:AT3G43190 SUS4; SUS4; UDP-glycosyltransferase/ sucrose synthase/
transferase, transferring glycosyl groups (EC:2.4.1.13);
K00695 sucrose synthase [EC:2.4.1.13]
Length=808
Score = 34.3 bits (77), Expect = 0.087, Method: Composition-based stats.
Identities = 17/38 (44%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEP 38
FG+ EAM G A +GGP E IV G++GF +P
Sbjct 677 FGLTVVEAMTCGLPTFATCNGGPAEIIVHGKSGFHIDP 714
> ath:AT5G20830 SUS1; SUS1 (SUCROSE SYNTHASE 1); UDP-glycosyltransferase/
sucrose synthase (EC:2.4.1.13); K00695 sucrose synthase
[EC:2.4.1.13]
Length=808
Score = 34.3 bits (77), Expect = 0.11, Method: Composition-based stats.
Identities = 17/38 (44%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEP 38
FG+ EAM G A GGP E IV G++GF +P
Sbjct 677 FGLTVVEAMTCGLPTFATCKGGPAEIIVHGKSGFHIDP 714
> hsa:440138 ALG11, CDG1P, GT8, KIAA0266, UTP14C; asparagine-linked
glycosylation 11, alpha-1,2-mannosyltransferase homolog
(yeast) (EC:2.4.-.-); K03844 alpha-1,2-mannosyltransferase
[EC:2.4.1.-]
Length=492
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 27/77 (35%), Positives = 40/77 (51%), Gaps = 8/77 (10%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIV---DGE-TGFLCEPTPASFSAALQRILLLSRD 56
FGI E M G + +A NSGGP+ IV +G+ TGFL E + ++ + IL +
Sbjct 400 FGIGVVECMAAGTIILAHNSGGPKLDIVVPHEGDITGFLAE-SEEDYAETIAHILSM--- 455
Query 57 SPEELAALRSKAEAHAA 73
S E+ +R A A +
Sbjct 456 SAEKRLQIRKSARASVS 472
> dre:407614 alg11; asparagine-linked glycosylation 11, alpha-1,2-mannosyltransferase;
K03844 alpha-1,2-mannosyltransferase
[EC:2.4.1.-]
Length=500
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 23/58 (39%), Positives = 33/58 (56%), Gaps = 5/58 (8%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIV----DGETGFLCEPTPASFSAALQRILLLS 54
FGI E M G + +A SGGP+ IV G TGFL + +++ A++RIL +S
Sbjct 404 FGIGIVECMAAGTIILAHKSGGPKLDIVVPYDGGPTGFLADDED-NYADAMERILSMS 460
> eco:b2761 ygcB, cas3, ECK2756, JW2731; Cas3 predicted helicase
needed for Cascade anti-viral activity; K07012
Length=888
Score = 32.7 bits (73), Expect = 0.28, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 25/51 (49%), Gaps = 0/51 (0%)
Query 69 EAHAACCFSPDVFRRSLHGILEEAAGPSDEISLNAPEKFPTGDCRAQLKRR 119
E + A F PD +R+ L I ++A E N +KF + +C + K R
Sbjct 715 ELNGASLFFPDAYRQWLDSIYDDAEMDEPEWVGNGMDKFESAECEKRFKAR 765
> ath:AT1G32900 starch synthase, putative; K13679 granule-bound
starch synthase [EC:2.4.1.242]
Length=610
Score = 32.3 bits (72), Expect = 0.39, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 2 GIVPCEAMFIGGLPVACNSGGPRETIVDGETGF 34
G++ AM G +P+ ++GG +T+ DG TGF
Sbjct 489 GLIQLHAMRYGTVPIVASTGGLVDTVKDGYTGF 521
> xla:399219 thrap3, MGC132228, trap150; thyroid hormone receptor
associated protein 3; K13112 thyroid hormone receptor-associated
protein 3
Length=918
Score = 32.0 bits (71), Expect = 0.48, Method: Composition-based stats.
Identities = 24/82 (29%), Positives = 35/82 (42%), Gaps = 4/82 (4%)
Query 40 PASFSAALQRILLLSRDSPEELAALRSKAEAHAACCFSPDVFRRSLHGILEEAAGPSDEI 99
P+S + + + L R +E + + E H SP FR+ HG L E A S E
Sbjct 607 PSSMTLSERFAKYLKRAKDQESSKSKKSPEIHRRIDISPSAFRK--HGFLHEEAKHSKET 664
Query 100 SLNAPEKF--PTGDCRAQLKRR 119
L K+ D R ++RR
Sbjct 665 GLKGDGKYRDEPSDLRQDIERR 686
> hsa:6643 SNX2, MGC5204, TRG-9; sorting nexin 2
Length=519
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 32/108 (29%), Positives = 48/108 (44%), Gaps = 17/108 (15%)
Query 16 VACNSGGPRETIV---DGETGFLCEPTPASFSAALQRILLLSRD--------SPEELAAL 64
++ NS GP+ T V D E T + +R +LS + +P L A
Sbjct 52 ISANSNGPKPTEVVLDDDREDLFAEATEEVSLDSPEREPILSSEPSPAVTPVTPTTLIAP 111
Query 65 RSKAEAHAACCFSPDVFRRSLHGILEEAAGP--SDEISLNAPEKFPTG 110
R ++++ +A P +F RS I EEA G EI ++ PEK G
Sbjct 112 RIESKSMSA----PVIFDRSREEIEEEANGDIFDIEIGVSDPEKVGDG 155
> ath:AT1G67623 F-box family protein
Length=296
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 37 EPTPASFSAALQRILLLSRDSPEELAALRSKAEAHAACCFSPDVFRRS 84
+ T FS+A + L ++R+ P E + E + AC F P+ F R+
Sbjct 226 DETDLDFSSACELYLYINRNFPFEAKKSKIDGEIYWACEFEPNRFVRT 273
> ath:AT4G02280 SUS3; SUS3 (sucrose synthase 3); UDP-glycosyltransferase/
sucrose synthase/ transferase, transferring glycosyl
groups (EC:2.4.1.13); K00695 sucrose synthase [EC:2.4.1.13]
Length=809
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEP 38
FG+ EAM G A GGP E I G +GF +P
Sbjct 678 FGLTVVEAMTCGLPTFATCHGGPAEIIEHGLSGFHIDP 715
> dre:407679 topbp1; topoisomerase (DNA) II binding protein 1
Length=1526
Score = 28.9 bits (63), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 33/85 (38%), Gaps = 6/85 (7%)
Query 2 GIVPCEAMFIGGLPVACN-----SGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRD 56
++ + F GG V N G R + G L +P+P+ F + SR
Sbjct 1392 NVIVVQGAF-GGWIVMLNIDQAREAGFRRLLQSGGAKVLPDPSPSLFKETTHLFVDFSRL 1450
Query 57 SPEELAALRSKAEAHAACCFSPDVF 81
P ++ S+A AH C P+
Sbjct 1451 KPGDVRVDISEASAHGVKCLKPEYI 1475
> cel:F53B7.5 hypothetical protein
Length=3095
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 15 PVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSP 58
P+ P TI+ G+T +L P A+ + ++ +LLS SP
Sbjct 672 PIEIGDTTPVSTILSGDTSYLSAPLSANLACRDKKWVLLSSTSP 715
> dre:565516 hypothetical LOC565516
Length=634
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 10/21 (47%), Positives = 14/21 (66%), Gaps = 0/21 (0%)
Query 22 GPRETIVDGETGFLCEPTPAS 42
GP I+DG +G+LCE P +
Sbjct 461 GPHTRIIDGGSGYLCEMEPVA 481
> ath:AT5G40100 disease resistance protein (TIR-NBS-LRR class),
putative
Length=1017
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 14/36 (38%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 48 QRILLLSRDSPEELAALRSKAEAHAACCFSPDVFRR 83
+ I+ + + S E LA A H AC F+ D FRR
Sbjct 416 ENIMEILKISYEGLAKAHQNAFLHVACLFNGDTFRR 451
> tgo:TGME49_046980 hypothetical protein
Length=2085
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 46/111 (41%), Gaps = 13/111 (11%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FGI + + G VA NSGGPR+ I+ + G E + S+ + D+P
Sbjct 885 FGIAVVQLLCAGCCVVAHNSGGPRDDILVAKRGEKSEESANVESSQV--------DAPHA 936
Query 61 LAALRSKAEAHAACCFSPDVFRRSLHGILEEAAGPSDEI---SLNAPEKFP 108
L AL + E + C F ++ +L E S NA EK P
Sbjct 937 L-ALTCRGE-YGFLCSDETEFAATIAAVLSETTDSCRHFLPDSRNAAEKTP 985
> xla:380176 snx2, MGC64590; sorting nexin 2
Length=519
Score = 28.1 bits (61), Expect = 7.1, Method: Composition-based stats.
Identities = 30/108 (27%), Positives = 47/108 (43%), Gaps = 17/108 (15%)
Query 16 VACNSGGPRET---IVDGETGFLCEPTPASFSAALQRILLLSRD--------SPEELAAL 64
++ NS GP+ T + D E T + +R L+LS + +P L
Sbjct 52 ISTNSNGPKTTEVLLCDDREDLFAEATDEVSLDSPERDLILSSEPSPAITPVTPTSLITP 111
Query 65 RSKAEAHAACCFSPDVFRRSLHGILEEAAGPSD--EISLNAPEKFPTG 110
R + + +A P ++ RS I EEA G + EI ++ PEK G
Sbjct 112 RMETISISA----PVIYDRSTDEIEEEANGDAFDLEIGVSDPEKVGDG 155
> ath:AT5G01220 SQD2; SQD2 (sulfoquinovosyldiacylglycerol 2);
UDP-glycosyltransferase/ UDP-sulfoquinovose:DAG sulfoquinovosyltransferase/
transferase, transferring glycosyl groups; K06119
sulfoquinovosyltransferase [EC:2.4.1.-]
Length=510
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 35/75 (46%), Gaps = 4/75 (5%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETI---VDGETGFLCEPTPASFSAALQRILLLSRDS 57
G+V EAM G VA +GG + I +G+TGFL P R LL R++
Sbjct 389 LGLVVLEAMSSGLPVVAARAGGIPDIIPEDQEGKTGFLFNPGDVEDCVTKLRTLLHDRET 448
Query 58 PEELA-ALRSKAEAH 71
E + A R + E +
Sbjct 449 REIIGKAAREETEKY 463
> ath:AT1G52420 glycosyl transferase family 1 protein
Length=670
Score = 27.7 bits (60), Expect = 8.5, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 26/61 (42%), Gaps = 0/61 (0%)
Query 1 FGIVPCEAMFIGGLPVACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPEE 60
FG V EAM G V ++GG +E + TG L + +L L R+ E
Sbjct 577 FGRVTIEAMAYGLAVVGTDAGGTKEMVQHNMTGLLHSMGRSGNKELAHNLLYLLRNPDER 636
Query 61 L 61
L
Sbjct 637 L 637
> ath:AT3G15940 glycosyl transferase family 1 protein
Length=697
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 28/62 (45%), Gaps = 2/62 (3%)
Query 1 FGIVPCEAMFIGGLPV-ACNSGGPRETIVDGETGFLCEPTPASFSAALQRILLLSRDSPE 59
FG V EAM GLPV ++GG +E + TG L A Q +L L R+
Sbjct 604 FGRVTIEAMAY-GLPVLGTDAGGTKEIVEHNVTGLLHPVGRAGNKVLAQNLLFLLRNPST 662
Query 60 EL 61
L
Sbjct 663 RL 664
Lambda K H
0.320 0.137 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2013067560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40