bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4384_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
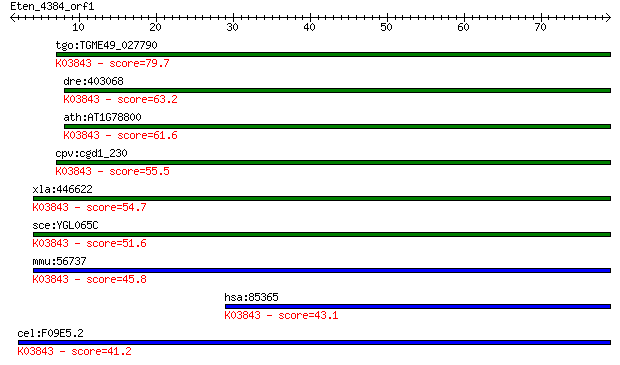
tgo:TGME49_027790 glycosyl transferase, group 1 domain contain... 79.7 2e-15
dre:403068 alg2, FLJ14511, im:7145131; asparagine-linked glyco... 63.2 2e-10
ath:AT1G78800 glycosyl transferase family 1 protein; K03843 al... 61.6 6e-10
cpv:cgd1_230 ALG-2 like alpha-1,3 mannosyltransferase ; K03843... 55.5 4e-08
xla:446622 alg2, MGC82301; asparagine-linked glycosylation 2 h... 54.7 8e-08
sce:YGL065C ALG2; Alg2p (EC:2.4.1.132 2.4.1.-); K03843 alpha-1... 51.6 6e-07
mmu:56737 Alg2, 1110018A23Rik, 1300013N08Rik, ALPG2, CDGIi, MN... 45.8 4e-05
hsa:85365 ALG2, CDGIi, FLJ14511, NET38, hALPG2; asparagine-lin... 43.1 2e-04
cel:F09E5.2 hypothetical protein; K03843 alpha-1,3/alpha-1,6-m... 41.2 7e-04
> tgo:TGME49_027790 glycosyl transferase, group 1 domain containing
protein (EC:2.4.1.14); K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=506
Score = 79.7 bits (195), Expect = 2e-15, Method: Composition-based stats.
Identities = 36/72 (50%), Positives = 47/72 (65%), Gaps = 0/72 (0%)
Query 7 RLNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVNPLLRL 66
RL + +G FLP SI FCS+LRML+LVL +TG +++ NDQVA VNPLL
Sbjct 106 RLKVFEYGHFLPRSIFGKAVAFCSLLRMLWLVLVIFVTGRWRRNVIINDQVAAVNPLLSF 165
Query 67 VGKKVIFYGHFP 78
+K++FY HFP
Sbjct 166 FCEKLVFYAHFP 177
> dre:403068 alg2, FLJ14511, im:7145131; asparagine-linked glycosylation
2 homolog (S. cerevisiae, alpha-1,3-mannosyltransferase);
K03843 alpha-1,3/alpha-1,6-mannosyltransferase [EC:2.4.1.132
2.4.1.-]
Length=402
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 31/73 (42%), Positives = 44/73 (60%), Gaps = 2/73 (2%)
Query 8 LNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVNPLLRLV 67
L +V G +LP S+ +F C+ LRM+YL + ++ +D+VF DQV+ P LRL
Sbjct 55 LPVVCVGDWLPTSVFGYFHALCAYLRMIYLTIYLVLFSGEEFDVVFCDQVSACIPFLRLA 114
Query 68 --GKKVIFYGHFP 78
KKV+FY HFP
Sbjct 115 RQRKKVLFYCHFP 127
> ath:AT1G78800 glycosyl transferase family 1 protein; K03843
alpha-1,3/alpha-1,6-mannosyltransferase [EC:2.4.1.132 2.4.1.-]
Length=403
Score = 61.6 bits (148), Expect = 6e-10, Method: Composition-based stats.
Identities = 31/72 (43%), Positives = 43/72 (59%), Gaps = 2/72 (2%)
Query 8 LNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVNPLLRLV 67
+ V+GSFLP I C+ LR L++ L ++ G +D+V DQV+VV PLL+L
Sbjct 62 FQVTVYGSFLPRHIFYRLHAVCAYLRCLFVAL-CVLLGWSSFDVVLADQVSVVVPLLKLK 120
Query 68 -GKKVIFYGHFP 78
KV+FY HFP
Sbjct 121 RSSKVVFYCHFP 132
> cpv:cgd1_230 ALG-2 like alpha-1,3 mannosyltransferase ; K03843
alpha-1,3/alpha-1,6-mannosyltransferase [EC:2.4.1.132 2.4.1.-]
Length=474
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 27/82 (32%), Positives = 49/82 (59%), Gaps = 10/82 (12%)
Query 7 RLNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLT----AIMTGHV------GYDLVFNDQ 56
R+ + V+G+++P + + S +R++Y L +MT YD++ NDQ
Sbjct 73 RIKVKVYGNWIPRTFFGYGTTLFSYIRIIYTSLIMFFFVMMTSFSLEKTSRYYDVILNDQ 132
Query 57 VAVVNPLLRLVGKKVIFYGHFP 78
V+V+NP+L+L+ +++IFY HFP
Sbjct 133 VSVINPILKLMTRRLIFYCHFP 154
> xla:446622 alg2, MGC82301; asparagine-linked glycosylation 2
homolog; K03843 alpha-1,3/alpha-1,6-mannosyltransferase [EC:2.4.1.132
2.4.1.-]
Length=404
Score = 54.7 bits (130), Expect = 8e-08, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 42/77 (54%), Gaps = 2/77 (2%)
Query 4 KTSRLNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVNPL 63
+ S + I G +LP S+L C+ +RM++L L + +D+VF DQV+ P
Sbjct 52 RNSGIPIRCCGDWLPRSLLGRCHALCAYIRMIFLALYIVFLSGEQFDVVFCDQVSACIPF 111
Query 64 LRLV--GKKVIFYGHFP 78
+L KKV+FY HFP
Sbjct 112 FKLARNSKKVLFYCHFP 128
> sce:YGL065C ALG2; Alg2p (EC:2.4.1.132 2.4.1.-); K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=503
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 43/77 (55%), Gaps = 2/77 (2%)
Query 4 KTSRLNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHV-GYDLVFNDQVAVVNP 62
K +L + V+G FLP + L F + + +R LYLV+ I+ V Y L+ DQ++ P
Sbjct 56 KNGQLKVEVYGDFLPTNFLGRFFIVFATIRQLYLVIQLILQKKVNAYQLIIIDQLSTCIP 115
Query 63 LLRLVGK-KVIFYGHFP 78
LL + ++FY HFP
Sbjct 116 LLHIFSSATLMFYCHFP 132
> mmu:56737 Alg2, 1110018A23Rik, 1300013N08Rik, ALPG2, CDGIi,
MNCb-5081; asparagine-linked glycosylation 2 homolog (yeast,
alpha-1,3-mannosyltransferase); K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=415
Score = 45.8 bits (107), Expect = 4e-05, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 43/79 (54%), Gaps = 4/79 (5%)
Query 4 KTSRLNIVVFGSFLPHSILRHFR--VFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVN 61
+T L++ G +LP S+ R CS +RM++L L + +D+V DQV+
Sbjct 62 ETRELSVQCAGDWLPRSLGWGGRGAAICSYVRMVFLALYVLFLSGEEFDVVVCDQVSACI 121
Query 62 PLLRLV--GKKVIFYGHFP 78
P+ +L K+V+FY HFP
Sbjct 122 PVFKLARRRKRVLFYCHFP 140
> hsa:85365 ALG2, CDGIi, FLJ14511, NET38, hALPG2; asparagine-linked
glycosylation 2, alpha-1,3-mannosyltransferase homolog
(S. cerevisiae) (EC:2.4.1.-); K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=416
Score = 43.1 bits (100), Expect = 2e-04, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 31/52 (59%), Gaps = 2/52 (3%)
Query 29 CSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVNPLLRLV--GKKVIFYGHFP 78
C+ +RM++L L + +D+V DQV+ P+ RL KK++FY HFP
Sbjct 89 CAYVRMVFLALYVLFLADEEFDVVVCDQVSACIPVFRLARRRKKILFYCHFP 140
> cel:F09E5.2 hypothetical protein; K03843 alpha-1,3/alpha-1,6-mannosyltransferase
[EC:2.4.1.132 2.4.1.-]
Length=400
Score = 41.2 bits (95), Expect = 7e-04, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 2/78 (2%)
Query 2 LQKTSRLNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVN 61
Q+T L+I ++P SI C+ L+M+ L I+ H D++ +D V+
Sbjct 47 FQETLDLDICTVVPWIPRSIFGKGHALCAYLKMIIAALY-IVIYHKDTDVILSDSVSASQ 105
Query 62 PLLRLVGK-KVIFYGHFP 78
+LR K K++FY HFP
Sbjct 106 FVLRHFSKAKLVFYCHFP 123
Lambda K H
0.336 0.149 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2067704464
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40