bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4347_orf1
Length=100
Score E
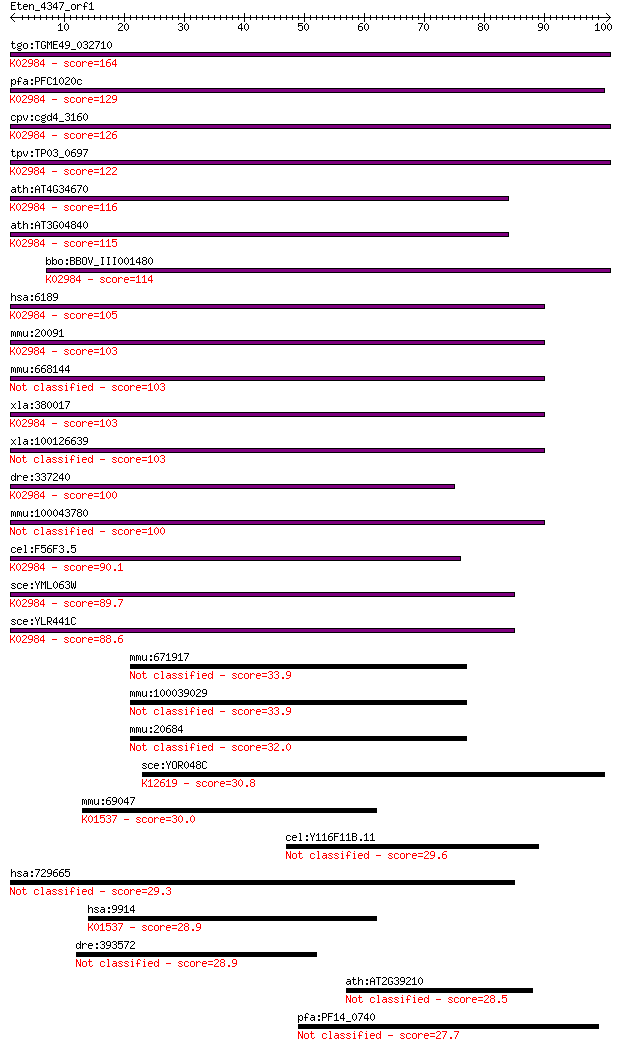
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_032710 40S ribosomal protein S3a, putative ; K02984... 164 4e-41
pfa:PFC1020c 40S ribosomal protein S3A, putative; K02984 small... 129 2e-30
cpv:cgd4_3160 40S ribosomal protein S3A ; K02984 small subunit... 126 1e-29
tpv:TP03_0697 40S ribosomal protein S3A; K02984 small subunit ... 122 3e-28
ath:AT4G34670 40S ribosomal protein S3A (RPS3aB); K02984 small... 116 2e-26
ath:AT3G04840 40S ribosomal protein S3A (RPS3aA); K02984 small... 115 3e-26
bbo:BBOV_III001480 17.m07150; 40S ribosomal protein S3Ae; K029... 114 9e-26
hsa:6189 RPS3A, FTE1, MFTL, MGC23240; ribosomal protein S3A; K... 105 3e-23
mmu:20091 Rps3a, MGC102469, MGC117914; ribosomal protein S3A; ... 103 9e-23
mmu:668144 Gm9000, EG668144; predicted gene 9000 103 9e-23
xla:380017 rps3a-a, MGC53968, fte1, mftl, rps3a; ribosomal pro... 103 1e-22
xla:100126639 rps3a-b, fte1, mftl; ribosomal protein S3A 103 2e-22
dre:337240 rps3a, MGC86672, fb02h01, rpS3Ae, wu:fb02h01, zgc:7... 100 9e-22
mmu:100043780 Gm10119; predicted gene 10119 100 1e-21
cel:F56F3.5 rps-1; Ribosomal Protein, Small subunit family mem... 90.1 1e-18
sce:YML063W RPS1B, PLC2, RP10B; Rps1bp; K02984 small subunit r... 89.7 2e-18
sce:YLR441C RPS1A, RP10A; Rps1ap; K02984 small subunit ribosom... 88.6 4e-18
mmu:671917 component of Sp100-rs-like 33.9 0.12
mmu:100039029 component of Sp100-rs-like 33.9 0.13
mmu:20684 Sp100, A430075G10Rik; nuclear antigen Sp100 32.0 0.56
sce:YOR048C RAT1, HKE1, TAP1, XRN2; Nuclear 5' to 3' single-st... 30.8 0.96
mmu:69047 Atp2c2, 1810010G06Rik, KIAA0703, Spca2, mKIAA0703; A... 30.0 1.6
cel:Y116F11B.11 hypothetical protein 29.6 2.3
hsa:729665 C14orf38, c14_5395; chromosome 14 open reading fram... 29.3 3.3
hsa:9914 ATP2C2, DKFZp686H22230, KIAA0703, SPCA2; ATPase, Ca++... 28.9 3.7
dre:393572 traf3ip1, Elipsa, MGC63522, zgc:63522; TNF receptor... 28.9 4.1
ath:AT2G39210 nodulin family protein 28.5 5.9
pfa:PF14_0740 Plasmodium exported protein (hyp17), unknown fun... 27.7 9.8
> tgo:TGME49_032710 40S ribosomal protein S3a, putative ; K02984
small subunit ribosomal protein S3Ae
Length=259
Score = 164 bits (416), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 80/100 (80%), Positives = 90/100 (90%), Gaps = 1/100 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
IRAIRKKM++IM+AEASKCQLRDLVKKFIPESIGK IE AC+ IFPLQNV IRKVK++KK
Sbjct 161 IRAIRKKMMTIMSAEASKCQLRDLVKKFIPESIGKDIENACKGIFPLQNVFIRKVKMLKK 220
Query 61 PKFDLSKLMELHGETSEDVGKKVATESAAAQNTLSAEVKA 100
PKFDL+KLMELHGE +EDVG+KV ES AQNTL+AE KA
Sbjct 221 PKFDLTKLMELHGE-AEDVGRKVQAESGEAQNTLTAETKA 259
> pfa:PFC1020c 40S ribosomal protein S3A, putative; K02984 small
subunit ribosomal protein S3Ae
Length=262
Score = 129 bits (324), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 66/101 (65%), Positives = 78/101 (77%), Gaps = 2/101 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
I+ IRKKMV IM AEASK L+DLVKKFIPESIGK IE C+ I+PLQNVLIRKVK++K+
Sbjct 161 IKKIRKKMVDIMTAEASKVLLKDLVKKFIPESIGKEIEKQCKKIYPLQNVLIRKVKILKR 220
Query 61 PKFDLSKLMELHGETSEDVGKKVAT--ESAAAQNTLSAEVK 99
PK D+SKLMELH + E+ GK V ES A N L+AE+K
Sbjct 221 PKLDISKLMELHTDPKEESGKNVNALPESKEATNILTAELK 261
> cpv:cgd4_3160 40S ribosomal protein S3A ; K02984 small subunit
ribosomal protein S3Ae
Length=262
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 66/102 (64%), Positives = 76/102 (74%), Gaps = 2/102 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
IR IRKKMV I+ AEA LRDLVKKFIPESIGK IE ACR IFPLQ+V IRKVKV+ K
Sbjct 161 IRQIRKKMVEIITAEAEGTTLRDLVKKFIPESIGKEIEKACRHIFPLQHVFIRKVKVLSK 220
Query 61 PKFDLSKLMELH--GETSEDVGKKVATESAAAQNTLSAEVKA 100
P FD+S+LME H + E+ G KV +ES A N L+AE+KA
Sbjct 221 PSFDVSRLMESHVGADGVEEKGTKVVSESNQATNLLTAEMKA 262
> tpv:TP03_0697 40S ribosomal protein S3A; K02984 small subunit
ribosomal protein S3Ae
Length=262
Score = 122 bits (306), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 60/101 (59%), Positives = 78/101 (77%), Gaps = 1/101 (0%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
+R I KKMVS+M EAS L++LVKK IPESIGK IE AC+SIFPLQNVL+RK+KV+KK
Sbjct 161 VRLIHKKMVSVMTNEASNSTLKELVKKVIPESIGKEIEKACKSIFPLQNVLVRKIKVLKK 220
Query 61 PKFDLSKLMELHGETSEDVGKKVAT-ESAAAQNTLSAEVKA 100
PKFDL+KLME H + ++ G+ + ES A N L+AE+++
Sbjct 221 PKFDLTKLMESHNYSGDEEGRTLKVKESENATNLLTAELQS 261
> ath:AT4G34670 40S ribosomal protein S3A (RPS3aB); K02984 small
subunit ribosomal protein S3Ae
Length=262
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 55/84 (65%), Positives = 67/84 (79%), Gaps = 1/84 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
IR IR+KM IM EAS C L++LV KFIPE+IG+ IE A + I+PLQNV IRKVK++K
Sbjct 161 IRQIRRKMSEIMVKEASSCDLKELVAKFIPEAIGREIEKATQGIYPLQNVFIRKVKILKA 220
Query 61 PKFDLSKLMELHGE-TSEDVGKKV 83
PKFDL KLME+HG+ T+EDVG KV
Sbjct 221 PKFDLGKLMEVHGDYTAEDVGVKV 244
> ath:AT3G04840 40S ribosomal protein S3A (RPS3aA); K02984 small
subunit ribosomal protein S3Ae
Length=262
Score = 115 bits (288), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 55/84 (65%), Positives = 66/84 (78%), Gaps = 1/84 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
IR IR+KM IM EAS C L+DLV KFIPE+IG+ IE A + I+PLQNV IRKVK++K
Sbjct 161 IRQIRRKMRDIMVREASSCDLKDLVAKFIPEAIGREIEKATQGIYPLQNVFIRKVKILKA 220
Query 61 PKFDLSKLMELHGETS-EDVGKKV 83
PKFDL KLM++HG+ S EDVG KV
Sbjct 221 PKFDLGKLMDVHGDYSAEDVGVKV 244
> bbo:BBOV_III001480 17.m07150; 40S ribosomal protein S3Ae; K02984
small subunit ribosomal protein S3Ae
Length=264
Score = 114 bits (284), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 56/95 (58%), Positives = 71/95 (74%), Gaps = 1/95 (1%)
Query 7 KMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKKPKFDLS 66
KM+ +M EAS +R++VKK IPESIGK IE AC+S+FPLQNVLIRK+KV+KKPKFDL+
Sbjct 167 KMIEVMQREASTTSMREVVKKIIPESIGKEIEKACKSVFPLQNVLIRKIKVLKKPKFDLT 226
Query 67 KLMELHGETSEDVGKKV-ATESAAAQNTLSAEVKA 100
KLME+H + SE+ G ES A N L+ E+ A
Sbjct 227 KLMEVHTDISEETGHATKVEESEEAANLLTKELDA 261
> hsa:6189 RPS3A, FTE1, MFTL, MGC23240; ribosomal protein S3A;
K02984 small subunit ribosomal protein S3Ae
Length=264
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 50/89 (56%), Positives = 64/89 (71%), Gaps = 1/89 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
+R IRKKM+ IM E L+++V K IP+SIGK IE AC+SI+PL +V +RKVK++KK
Sbjct 161 VRQIRKKMMEIMTREVQTNDLKEVVNKLIPDSIGKDIEKACQSIYPLHDVFVRKVKMLKK 220
Query 61 PKFDLSKLMELHGETSEDVGKKVATESAA 89
PKF+L KLMELHGE S GK E+ A
Sbjct 221 PKFELGKLMELHGEGSSS-GKATGDETGA 248
> mmu:20091 Rps3a, MGC102469, MGC117914; ribosomal protein S3A;
K02984 small subunit ribosomal protein S3Ae
Length=264
Score = 103 bits (258), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 49/89 (55%), Positives = 63/89 (70%), Gaps = 1/89 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
+R IRKKM+ IM E L+++V K IP+SIGK IE AC+SI+PL +V +RKVK++KK
Sbjct 161 VRQIRKKMMEIMTREVQTNDLKEVVNKLIPDSIGKDIEKACQSIYPLHDVFVRKVKMLKK 220
Query 61 PKFDLSKLMELHGETSEDVGKKVATESAA 89
PKF+L KLMELHGE GK E+ A
Sbjct 221 PKFELGKLMELHGEGGSS-GKAAGDETGA 248
> mmu:668144 Gm9000, EG668144; predicted gene 9000
Length=264
Score = 103 bits (258), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 49/89 (55%), Positives = 63/89 (70%), Gaps = 1/89 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
+R IRKKM+ IM E L+++V K IP+SIGK IE AC+SI+PL +V +RKVK++KK
Sbjct 161 VRQIRKKMMEIMTREVQTNDLKEVVNKLIPDSIGKDIEKACQSIYPLHDVFVRKVKMLKK 220
Query 61 PKFDLSKLMELHGETSEDVGKKVATESAA 89
PKF+L KLMELHGE GK E+ A
Sbjct 221 PKFELGKLMELHGEGGSS-GKAAGDETGA 248
> xla:380017 rps3a-a, MGC53968, fte1, mftl, rps3a; ribosomal protein
S3A; K02984 small subunit ribosomal protein S3Ae
Length=264
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 49/89 (55%), Positives = 62/89 (69%), Gaps = 1/89 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
+R IRKKM IM E L+++V K IP+SIGK IE AC+SI+PL +V +RKVK++KK
Sbjct 161 VRQIRKKMFEIMTREVQTNDLKEVVNKLIPDSIGKDIEKACQSIYPLHDVYVRKVKMLKK 220
Query 61 PKFDLSKLMELHGETSEDVGKKVATESAA 89
PKF+L KLMELHGE GK E+ A
Sbjct 221 PKFELGKLMELHGEGG-GTGKPAGDETGA 248
> xla:100126639 rps3a-b, fte1, mftl; ribosomal protein S3A
Length=264
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 50/89 (56%), Positives = 64/89 (71%), Gaps = 1/89 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
+R IRKKM+ IM+ E L+++V K IP+SIGK IE AC+SI+PL +V RKVK++KK
Sbjct 161 VRQIRKKMMEIMSREVQTNDLKEVVNKLIPDSIGKDIEKACQSIYPLHDVYARKVKMLKK 220
Query 61 PKFDLSKLMELHGETSEDVGKKVATESAA 89
PKF+L KLMELHGE GK A E+ A
Sbjct 221 PKFELGKLMELHGEGGGS-GKPAADETGA 248
> dre:337240 rps3a, MGC86672, fb02h01, rpS3Ae, wu:fb02h01, zgc:73195,
zgc:86672; ribosomal protein S3A; K02984 small subunit
ribosomal protein S3Ae
Length=267
Score = 100 bits (250), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 44/74 (59%), Positives = 58/74 (78%), Gaps = 0/74 (0%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
+R IRKKM+ IM E L+++V K IP+S+GK IE AC+SI+PL +V +RKVK++KK
Sbjct 161 VRQIRKKMMEIMTREVQTNDLKEVVNKLIPDSVGKDIEKACQSIYPLHDVYVRKVKMLKK 220
Query 61 PKFDLSKLMELHGE 74
PKF+L KLMELHGE
Sbjct 221 PKFELGKLMELHGE 234
> mmu:100043780 Gm10119; predicted gene 10119
Length=264
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 48/89 (53%), Positives = 62/89 (69%), Gaps = 1/89 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
+R IRKKM+ IM E L+++V K IP+SIGK IE AC+SI+PL +V +RKVK++KK
Sbjct 161 VRQIRKKMMEIMTREVQTNDLKEVVNKLIPDSIGKDIEKACQSIYPLHDVFVRKVKMLKK 220
Query 61 PKFDLSKLMELHGETSEDVGKKVATESAA 89
PK +L KLMELHGE GK E+ A
Sbjct 221 PKSELGKLMELHGEGGSS-GKAAGDETGA 248
> cel:F56F3.5 rps-1; Ribosomal Protein, Small subunit family member
(rps-1); K02984 small subunit ribosomal protein S3Ae
Length=257
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 38/75 (50%), Positives = 54/75 (72%), Gaps = 0/75 (0%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
IR IR +M+ + E + C L+++V K IP+SIGK IE C ++PLQ V IRKVK+IK+
Sbjct 159 IRKIRSEMIGCIEKEVTGCDLKEVVSKLIPDSIGKDIEKTCSKLYPLQEVYIRKVKIIKR 218
Query 61 PKFDLSKLMELHGET 75
PK DL +L +LHG++
Sbjct 219 PKVDLGRLHDLHGDS 233
> sce:YML063W RPS1B, PLC2, RP10B; Rps1bp; K02984 small subunit
ribosomal protein S3Ae
Length=255
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 46/85 (54%), Positives = 57/85 (67%), Gaps = 1/85 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
IRAIRK + I+ E L L K IPE I K IE A + IFPLQN+ +RKVK++K+
Sbjct 161 IRAIRKVISEILTREVQNSTLAQLTSKLIPEVINKEIENATKDIFPLQNIHVRKVKLLKQ 220
Query 61 PKFDLSKLMELHGETS-EDVGKKVA 84
PKFD+ LM LHGE S E+ GKKV+
Sbjct 221 PKFDVGALMALHGEGSGEEKGKKVS 245
> sce:YLR441C RPS1A, RP10A; Rps1ap; K02984 small subunit ribosomal
protein S3Ae
Length=255
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 46/85 (54%), Positives = 56/85 (65%), Gaps = 1/85 (1%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKK 60
IRAIRK + I+ E L L K IPE I K IE A + IFPLQN+ +RKVK++K+
Sbjct 161 IRAIRKVISEILTKEVQGSTLAQLTSKLIPEVINKEIENATKDIFPLQNIHVRKVKLLKQ 220
Query 61 PKFDLSKLMELHGETS-EDVGKKVA 84
PKFD+ LM LHGE S E+ GKKV
Sbjct 221 PKFDVGALMALHGEGSGEEKGKKVT 245
> mmu:671917 component of Sp100-rs-like
Length=178
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Query 21 LRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKKPKFDLSKLMELHGETS 76
LRD ++FI + + + +CRS+ P+ V+ R ++ ++K KFD++ L EL E +
Sbjct 102 LRD--REFITGKMYEDLLDSCRSLVPVDKVIYRALEELEK-KFDMTVLCELFNEVN 154
> mmu:100039029 component of Sp100-rs-like
Length=178
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Query 21 LRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKKPKFDLSKLMELHGETS 76
LRD ++FI + + + +CRS+ P+ V+ R ++ ++K KFD++ L EL E +
Sbjct 102 LRD--REFITGKMYEDLLDSCRSLVPVDKVIYRALEELEK-KFDMTVLCELFNEVN 154
> mmu:20684 Sp100, A430075G10Rik; nuclear antigen Sp100
Length=591
Score = 32.0 bits (71), Expect = 0.56, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 34/56 (60%), Gaps = 3/56 (5%)
Query 21 LRDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKKPKFDLSKLMELHGETS 76
LRD +FI + + + +CRS+ P+ V+ R ++ ++K KFD++ L EL E +
Sbjct 50 LRD--HEFITGKMYEDLLDSCRSLVPVDKVIYRALEELEK-KFDMTVLCELFNEVN 102
> sce:YOR048C RAT1, HKE1, TAP1, XRN2; Nuclear 5' to 3' single-stranded
RNA exonuclease, involved in RNA metabolism, including
rRNA and snRNA processing as well as poly (A+) dependent
and independent mRNA transcription termination (EC:3.1.13.-);
K12619 5'-3' exoribonuclease 2 [EC:3.1.13.-]
Length=1006
Score = 30.8 bits (68), Expect = 0.96, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 41/77 (53%), Gaps = 5/77 (6%)
Query 23 DLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKKPKFDLSKLMELHGETSEDVGKK 82
+L+K E I ++ A ++ F L V+ K K+I K K L K E E ++D KK
Sbjct 481 ELMKDISKEEIDDAVSKANKTNFNLAEVM--KQKIINK-KHRLEKDNE-EEEIAKD-SKK 535
Query 83 VATESAAAQNTLSAEVK 99
V TE A ++ L AE+K
Sbjct 536 VKTEKAESECDLDAEIK 552
> mmu:69047 Atp2c2, 1810010G06Rik, KIAA0703, Spca2, mKIAA0703;
ATPase, Ca++ transporting, type 2C, member 2 (EC:3.6.3.8);
K01537 Ca2+-transporting ATPase [EC:3.6.3.8]
Length=944
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 29/51 (56%), Gaps = 6/51 (11%)
Query 13 AAEASKCQL--RDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKKP 61
+ E + C L ++++K F S+GK +EA C + N +IRK V+ +P
Sbjct 408 SGEGTVCLLPSKEVIKGFDNVSVGKLVEAGCVA----NNAVIRKNAVMGQP 454
> cel:Y116F11B.11 hypothetical protein
Length=511
Score = 29.6 bits (65), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 29/49 (59%), Gaps = 7/49 (14%)
Query 47 LQNVLIRKVKVIKK--PKFD-----LSKLMELHGETSEDVGKKVATESA 88
L+N+ I ++ I P+ D L KL+EL+G+T DV K+ A E+A
Sbjct 14 LKNIGIEDIEYINNIVPELDIVNNCLKKLLELNGKTLSDVEKQEAMETA 62
> hsa:729665 C14orf38, c14_5395; chromosome 14 open reading frame
38
Length=793
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 21/95 (22%), Positives = 47/95 (49%), Gaps = 11/95 (11%)
Query 1 IRAIRKKMVSIMAAEASKCQLRDLVKKFIPESIGKSIEAACR-----SIFPLQNVLIR-- 53
+ +RK + ++ E+ + + + +P SI + +E R ++F + + +R
Sbjct 81 LEEMRKATIDLLEIESMELNKLYYLLETLPNSIKRELEECVRDARRLNLFEINTIKMRIT 140
Query 54 ----KVKVIKKPKFDLSKLMELHGETSEDVGKKVA 84
+++++KK DL+K E GE E++ +K A
Sbjct 141 RTENEIELLKKKITDLTKYNEALGEKQEELARKHA 175
> hsa:9914 ATP2C2, DKFZp686H22230, KIAA0703, SPCA2; ATPase, Ca++
transporting, type 2C, member 2 (EC:3.6.3.8); K01537 Ca2+-transporting
ATPase [EC:3.6.3.8]
Length=946
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 29/50 (58%), Gaps = 6/50 (12%)
Query 14 AEASKCQL--RDLVKKFIPESIGKSIEAACRSIFPLQNVLIRKVKVIKKP 61
+ + C L ++++K+F S+GK +EA C + N +IRK V+ +P
Sbjct 411 GQGTVCLLPSKEVIKEFSNVSVGKLVEAGCVA----NNAVIRKNAVMGQP 456
> dre:393572 traf3ip1, Elipsa, MGC63522, zgc:63522; TNF receptor-associated
factor 3 interacting protein 1
Length=629
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 12 MAAEASKCQLRDLVKKFIPESIGKSIEAACRSIFPLQNVL 51
+ +EAS+ + R+LV + I E + SI+ CRS PL ++
Sbjct 504 LVSEASRKKERELVAREI-ERLRSSIQTVCRSALPLGKIM 542
> ath:AT2G39210 nodulin family protein
Length=601
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 14/31 (45%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 57 VIKKPKFDLSKLMELHGETSEDVGKKVATES 87
V +KPK D S+ + GE S++V +KV T S
Sbjct 289 VTEKPKLDSSEFKDDDGEESKEVVEKVKTPS 319
> pfa:PF14_0740 Plasmodium exported protein (hyp17), unknown function
Length=468
Score = 27.7 bits (60), Expect = 9.8, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Query 49 NVLIRKVKVIKKPKFDLSKLMELHGETSEDVGKKVATESAAAQNTLSAEV 98
NVL+RK +V+ + K D+SK E + ET D+ K+ T++ + A V
Sbjct 149 NVLLRKQRVLYEEKKDVSK--EGNSETLGDLRSKLKTKTEKTNRNIYANV 196
Lambda K H
0.316 0.128 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2041372988
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40