bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_4343_orf1
Length=94
Score E
Sequences producing significant alignments: (Bits) Value
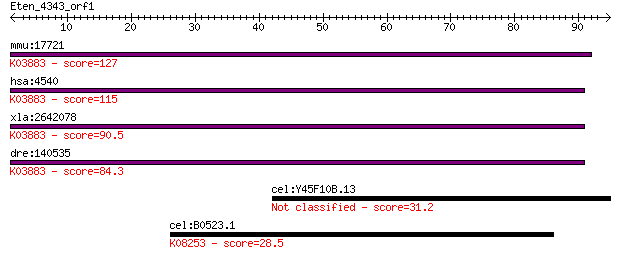
mmu:17721 ND5; NADH dehydrogenase subunit 5 (EC:1.6.5.3); K038... 127 1e-29
hsa:4540 ND5, MTND5; NADH dehydrogenase, subunit 5 (complex I)... 115 5e-26
xla:2642078 ND5; NADH dehydrogenase subunit 5; K03883 NADH deh... 90.5 1e-18
dre:140535 ND5, mtnd5; NADH dehydrogenase subunit 5; K03883 NA... 84.3 1e-16
cel:Y45F10B.13 hypothetical protein 31.2 0.83
cel:B0523.1 kin-31; protein KINase family member (kin-31); K08... 28.5 5.1
> mmu:17721 ND5; NADH dehydrogenase subunit 5 (EC:1.6.5.3); K03883
NADH dehydrogenase I subunit 5 [EC:1.6.5.3]
Length=607
Score = 127 bits (319), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 61/91 (67%), Positives = 79/91 (86%), Gaps = 0/91 (0%)
Query 1 ALLITLIATSFTAIYSTRIIFFALLGQPRFPTLVNINENNPLLINSIKRLLIGSLFAGYI 60
ALLITLIATS TA+YS RII+F + +PRFP L++INEN+P L+N IKRL GS+FAG++
Sbjct 408 ALLITLIATSMTAMYSMRIIYFVTMTKPRFPPLISINENDPDLMNPIKRLAFGSIFAGFV 467
Query 61 ISNNIPPTTIPQITMPYYLKTTALIVTILGF 91
IS NIPPT+IP +TMP++LKTTALI+++LGF
Sbjct 468 ISYNIPPTSIPVLTMPWFLKTTALIISVLGF 498
> hsa:4540 ND5, MTND5; NADH dehydrogenase, subunit 5 (complex
I); K03883 NADH dehydrogenase I subunit 5 [EC:1.6.5.3]
Length=603
Score = 115 bits (287), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 61/90 (67%), Positives = 69/90 (76%), Gaps = 0/90 (0%)
Query 1 ALLITLIATSFTAIYSTRIIFFALLGQPRFPTLVNINENNPLLINSIKRLLIGSLFAGYI 60
AL ITLIATS T+ YSTR+I L GQPRFPTL NINENNP L+N IKRL GSLFAG++
Sbjct 408 ALSITLIATSLTSAYSTRMILLTLTGQPRFPTLTNINENNPTLLNPIKRLAAGSLFAGFL 467
Query 61 ISNNIPPTTIPQITMPYYLKTTALIVTILG 90
I+NNI P + Q T+P YLK TAL VT LG
Sbjct 468 ITNNISPASPFQTTIPLYLKLTALAVTFLG 497
> xla:2642078 ND5; NADH dehydrogenase subunit 5; K03883 NADH dehydrogenase
I subunit 5 [EC:1.6.5.3]
Length=604
Score = 90.5 bits (223), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 50/90 (55%), Positives = 63/90 (70%), Gaps = 0/90 (0%)
Query 1 ALLITLIATSFTAIYSTRIIFFALLGQPRFPTLVNINENNPLLINSIKRLLIGSLFAGYI 60
AL +TLIATSFTAIYS R+IFFA +G PR L INENN +IN IKRL GS+ AG +
Sbjct 409 ALTLTLIATSFTAIYSFRVIFFASMGHPRSNPLSPINENNKTVINPIKRLAWGSIVAGLL 468
Query 61 ISNNIPPTTIPQITMPYYLKTTALIVTILG 90
I++N+ P P +TMP K A+IV++ G
Sbjct 469 IASNMLPINSPIMTMPTLAKQAAIIVSVTG 498
> dre:140535 ND5, mtnd5; NADH dehydrogenase subunit 5; K03883
NADH dehydrogenase I subunit 5 [EC:1.6.5.3]
Length=606
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 47/90 (52%), Positives = 62/90 (68%), Gaps = 1/90 (1%)
Query 1 ALLITLIATSFTAIYSTRIIFFALLGQPRFPTLVNINENNPLLINSIKRLLIGSLFAGYI 60
AL +TLIATSFTA+YS R+I+ LG PR T I+EN+ + N+I+RL GS+ AG I
Sbjct 409 ALTLTLIATSFTAVYSFRMIYLVCLGSPRHKTYETIDENH-IPTNTIQRLAWGSIIAGLI 467
Query 61 ISNNIPPTTIPQITMPYYLKTTALIVTILG 90
IS + P P +TMP YLK A++VT+LG
Sbjct 468 ISYTMIPLKTPILTMPIYLKLAAILVTLLG 497
> cel:Y45F10B.13 hypothetical protein
Length=618
Score = 31.2 bits (69), Expect = 0.83, Method: Composition-based stats.
Identities = 14/53 (26%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 42 LLINSIKRLLIGSLFAGYIISNNIPPTTIPQITMPYYLKTTALIVTILGFKEP 94
L + + + + G ++A +I+SNN+P T+P PY ++ + F+ P
Sbjct 140 LFLWKVLKCVFGMIYA-FIVSNNVPSQTLPSTPKPYTATPSSPSYNKVSFEYP 191
> cel:B0523.1 kin-31; protein KINase family member (kin-31); K08253
non-specific protein-tyrosine kinase [EC:2.7.10.2]
Length=437
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query 26 GQPRFPTLVNINENNPLLINS-IKRLLIGSLFAGYIISNNIPPTTIPQITMPYYLKTTAL 84
G+PR ++++ NN L NS +K +I S+ Y ++NN+ TI Q+ Y T +
Sbjct 50 GEPR-SYILSVMFNNKLDENSSVKHFVINSVENKYFVNNNMSFNTIQQMLSHYQKSRTEI 108
Query 85 I 85
+
Sbjct 109 L 109
Lambda K H
0.327 0.144 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069995292
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40